ABSTRACT
- Two rod-shaped, Gram-positive, spore-forming, motile, and strictly anaerobic bacteria, FM7315T and FM7330T were isolated from Myeolchi-jeot, a traditional Korean fermented anchovy. Phylogenetic and phylogenomic analyses based on the 16S rRNA gene and genome sequences revealed that strains FM7315T and FM7330T represent novel species within the genus Haloimpatiens. The genome sizes of strains FM7315T and FM7330T were 3,052,517 bp and 4,194,114 bp, respectively, with G + C contents of 29.7 mol% and 28.0 mol%, respectively. Strain FM7315T exhibited growth at 20–37°C, 0–2% NaCl, and pH range of 5.0–8.0, whereas strain FM7330T grew at 25–45°C, 0–4% NaCl, and pH range of 5.0–9.0. Strain FM7315T contains C14:0, C16:0, C18:1 ω9c, Summed Feature 3 (C16:1 ω7c/C16:1 ω6c), and Summed Feature 8 (C18:1 ω7c/C18:1 ω6c) as major fatty acids, along with diphosphatidylglycerol, phosphatidylglycerol, glycolipid, two aminophospholipids, and five unidentified lipids. Strain FM7330T contains C16:0, C17:1 ω8c, and C18:1 ω9c as major fatty acids, along with diphosphatidylglycerol, two phosphatidylglycerols, four aminophospholipids, and six unidentified lipids. Based on their phenotypic, chemotaxonomic, and molecular characteristics, strains FM7315T and FM7330T represent two novel species of the genus Haloimpatiens, for which the names Haloimpatiens sporogenes sp. nov. (FM7315T = KCTC 25939T = JCM 37574T) and Haloimpatiens myeolchijeotgali sp. nov. (FM7330T = KCTC 25938T = JCM 37575T) have been proposed.
-
Keywords: Haloimpatiens, novel species, Myeolchi-jeot, strictly anaerobic
Introduction
The family Clostridiaceae encompasses a phylogenetically and phenotypically diverse group of anaerobic spore-forming bacteria. A substantial number of species have been validly published and novel taxa within this family continue to be identified through ongoing taxonomic efforts (Wiegel et al., 2006; Yang et al., 2024). The genus Haloimpatiens, which belongs to the family Clostridiaceae, was first proposed by Wu et al. (2016). As of this writing, the genus Haloimpatiens comprises two species, with one validly published species, Haloimpatiens lingqiaonensis isolated from paper mills (Wu et al., 2016), and one invalidly published species, “Haloimpatiens massiliensis” isolated from the gut of healthy infants (Anani et al., 2020; Parte et al., 2020). Members of the genus Haloimpatiens are strictly anaerobic, rod-shaped, Gram-positive, sensitive to salt stress, and motile with peritrichous flagella. H. lingqiaonensis, the type species of the genus Haloimpatiens, is distinguished by its cellular polar lipids, including diphosphatidylglycerol (DPG), phosphatidylglycerol (PG), several phospholipids, and glycolipids (GLs), as well as by its major cellular fatty acids, C14:0 and C16:0. Additionally, the absence of quinones and their reliance on fermentation metabolism results in the production of various fermentation products such as formic acid, acetic acid, ethanol, and lactic acid (Wu et al., 2016). In this study, two novel strains belonging to the genus Haloimpatiens, designated as Haloimpatiens sporogenes FM7315T and Haloimpatiens myeolchijeotgali FM7330T, were isolated from Myeolchi-jeot, a traditional Korean fermented anchovy, and their taxonomic characteristics were assessed using a polyphasic approach.
Materials and Methods
Isolation and deposition of strains
The strains FM7315T and FM7330T were isolated from Myeolchi-jeot, a traditional Korean fermented anchovy, under anaerobic conditions. Briefly, Myeolchi-jeot samples were purchased from a traditional market in Ganggyeong-eup (36°09'37.3"N, 127°00'56.9"E), Republic of Korea. Following this, all procedures were performed in an anaerobic chamber (COY Laboratory Products) filled with 90% N2, 5% CO2, and 5% H2. The Myeolchi-jeot samples were serially diluted in phosphate-buffered saline, spread onto Bifidobacterium Selective agar (BS; MB cell), and incubated at 30°C for 7 d. After incubation, colonies grown on BS agar were randomly selected based on their morphology and their 16S rRNA genes were PCR-amplified using the universal primers 27F and 1492R (Lane et al., 1991). PCR amplicons were sequenced using the universal primer sets 337F (5’-GACTCCTACGGGAGGCWGCAG-3’), 785F (5’-GGATTAGATACCCTGGTA-3’), and 800R (5’-TACCAGGGTATCTAATCC-3’) (Macrogen, Korea). The sequencing results were assembled into 16S rRNA gene sequences using BioEdit version 7.7.1 (Hall et al., 1999). These sequences were compared with those of all published type species on the EzBioCloud server (www.ezbiocloud.net/identify). Based on this, two novel strains, FM7330T and FM7315T, belonging to the family Clostridiaceae, were selected for further taxonomic characterization. Strains FM7330T and FM7315T were routinely cultured anaerobically on BS agar at 40°C and 30°C, respectively, for 2 d, and in BL broth (MB Cell) under oxygen-free conditions achieved by nitrogen purging. The strains were preserved at –80°C in a solution containing 10% glycerol (v/v) and 10% skim milk (w/v). Strains FM7315T and FM7330T were deposited in the Korean Collection for Type Cultures (KCTC) and Japan Collection of Microorganisms (JCM) under accession numbers KCTC 25939T and JCM 37574T for FM7315T, and KCTC 25938T and JCM 37575T for FM7330T. To compare genomic and phenotypic characteristics between strains FM7315T and FM7330T, the type strain Haloimpatiens lingqiaonensis KCTC 15321T was obtained from the KCTC and was cultivated at 30°C for 2 d on GS agar (KCTC medium no. 511), with the medium prepared according to the KCTC media information.
16S rRNA gene-based phylogenetic analysis
The 16S rRNA gene sequences of strains FM7315T and FM7330T with those of closely related type strains were aligned using the SILVA Incremental Aligner (SINA) version 1.2.12 (Pruesse et al., 2012). Phylogenetic trees were then constructed based on neighbor-joining (NJ) (Saitou and Nei, 1987), maximum likelihood (ML) (Felsenstein et al., 1981), and maximum parsimony (MP) (Fitch et al., 1971) algorithms, with 1,000 bootstrap replications, using MEGA11 software (Tamura et al., 2021).
Whole-genome sequencing and phylogenomic analysis
Genomic DNA of strains FM7315T, FM7330T, and KCTC 15321T was extracted from cells cultured in optimal media using the standard phenol-chloroform extraction method (Sambrook et al., 1989). The genomic DNA of strain FM7315T was sequenced using the Illumina NovaSeqTM 6000 and PacBio Sequel II systems (Macrogen, Korea). Sequencing reads were assembled using Microbial Genome Analysis software (version SMRT LINK_13.1.0.221970). The genomic DNA of strains FM7330T and KCTC 15321T were sequenced using the Oxford Nanopore MinION platform. Nanopore sequencing reads were de novo-assembled using Flye software (version 2.9.1) (Kolmogorov et al., 2019). The quality of the assembled genomes of the strains FM7315T, FM7330T, and KCTC 15321T was evaluated based on their completeness and contamination rates using CheckM (version 1.2.2) (Chklovski et al., 2023) with a marker set for the family Clostridiaceae. For phylogenomic analysis of strains FM7315T and FM7330T, as well as their closely related type strains, the nucleotide sequences of 92 bacterial core genes were obtained from each genome, concatenated, and aligned using the updated bacterial core gene pipeline version 2 (UBCG2) (Kim et al., 2021). The concatenated alignment was then used to infer a ML phylogenomic tree with 1,000 bootstrap replications using MEGA11 software (Tamura et al., 2021).
To determine the genus-level affiliation of strains FM7315T and FM7330T, genome-based analyses of average amino acid identity (AAI; https://github.com/endixk/ezaai) and percentage of conserved proteins (POCP; https://github.com/hoelzer/pocp) were performed using the genomes of strains FM7315T, FM7330T, and closely related type strains from the genera Haloimpatiens (H. lingqiaonensis KCTC 15321T), and Clostridium (C. butyricum DSM 10702T). For species-level comparisons, the average nucleotide identity (ANI) (Lee et al., 2016) and digital DNA-DNA hybridization (dDDH) values were calculated against the closest type strain, H. lingqiaonensis KCTC 15321T. The dDDH values were obtained using the Genome-to-Genome Distance Calculator (GGDC; https://ggdc.dsmz.de/) based on formula 2 with the recommended BLAST+ method (Meier-Kolthoff et al., 2013).
Genome annotation and analysis
The whole-genome sequences of strains FM7315T, FM7330T, and KCTC 15321T were annotated using the NCBI Prokaryotic Genome Automatic Annotation Pipeline version 6.7 (Tatusova et al., 2016). The complete genomes of the strains FM7315T, FM7330T, and KCTC 15321T were visualized using Proksee (Grant et al., 2023). Functional genes were predicted based on the protein-coding sequences in the KEGG database (Kanehisa and Goto, 2000). The carbohydrate-active enzyme (CAZy) genes in the genomes of strains FM7315T, FM7330T, and KCTC 15321T were identified using the dbCAN3 meta server (https://bcb.unl.edu/dbCAN2/) with default parameters.
Physiological and morphological analyses
Strains FM7315T and FM7330T were tested for their ability to grow on BS agar, Brain Heart Infusion agar (BHI; MB Cell), Yeast extract Peptone Dextrose agar (YPD; MB Cell), and Reasoner’s 2A agar (R2A; Difco) at 30°C for 7 d under anaerobic conditions. The growth conditions of strains FM7315T and FM7330T were evaluated on BS agar at various temperatures (4–50°C), and in BL broth with different NaCl concentrations (0–10%, w/v) and pH values (4.0–10.0), all tested for 7 d under anaerobic conditions. BL broth of pH below 8.0 and pH 8.0–9.5 were prepared using Na2HPO4/NaH2PO4 and Tris/HCl buffers, respectively (Gomori, 1995). After sterilization (121°C for 15 min), the pH values were adjusted again, if necessary. Growth of strains FM7315T and FM7330T under aerobic condition was evaluated on BS agar at their optimal growth temperature for 7 d. Cell morphology of strains FM7315T and FM7330T was examined using field emission scanning electron microscopy (FE-SEM; JSM-6700F, JEOL). Motility was determined using semi-solid BL agar (0.4% agar) by stab inoculation (Tittsler and Sandholzer, 1936). Gram staining was performed as described by Gram (1884), and spore formation was examined by endospore staining of heat-treated cells (80°C for 20 min) (Reynolds et al., 2009). The oxidase and catalase activities were tested using an oxidase reagent (bioMérieux) and 3% hydrogen peroxide (Junsei), respectively. Hydrolytic activities of strains FM7315T and FM7330T were evaluated on BS agar supplemented with 6% (w/v) skim milk, starch, Tween 20, and Tween 80 (Gerhardt et al., 1994). The substrate utilization profiles of strains FM7315T and FM7330T, diverse sugars (20 mM), alcohols (20 mM), organic acids (20 mM), and amino acids (10 mM), were evaluated in minimal basal medium (Hernández-Eugenio et al., 2002). Fermentation products of strains FM7315T and FM7330T were analyzed using culture supernatants obtained after 3 days of incubation in BL broth under optimal growth conditions. Organic acids and alcohols were subsequently analyzed using liquid chromatography–tandem mass spectrometry with triple quadrupole and gas chromatography–mass spectrometry, respectively, according to Agilent Technologies (2016) and Lee et al. (2013). Biochemical properties were assessed using an API ZYM kit (bioMérieux) according to the manufacturer’s instructions, with all procedures performed under strict anaerobic conditions.
Chemotaxonomic characterization
For cellular fatty acid analysis, strains FM7315T and FM7330T were cultivated on BL medium, while Haloimpatiens lingqiaonensis KCTC 15321T was grown on GS medium under their respective optimal growth conditions. Cellular fatty acids were extracted using the standard MIDI protocol (Sherlock Microbial Identification System, version 6.0B) and analyzed using a gas chromatograph (Hewlett-Packard 6890) with reference to the TSBA6 database (Sasser, 1990). For polar lipid analysis, strains FM7315T and FM7330T were cultivated on BL medium, under their respective optimal growth conditions. Polar lipids were extracted using the chloroform–methanol method and subsequently analyzed via two-dimensional thin-layer chromatography, as described by Minnikin et al. (1977). The polar lipid composition was determined using four spray reagents: 10% ethanolic molybdophosphoric acid for total polar lipids, ninhydrin for aminolipids, Dittmer-Lester reagent for phospholipids, and α-naphthol and sulfuric acid for GLs.
Results and Discussion
16S rRNA gene-based phylogenetic analysis
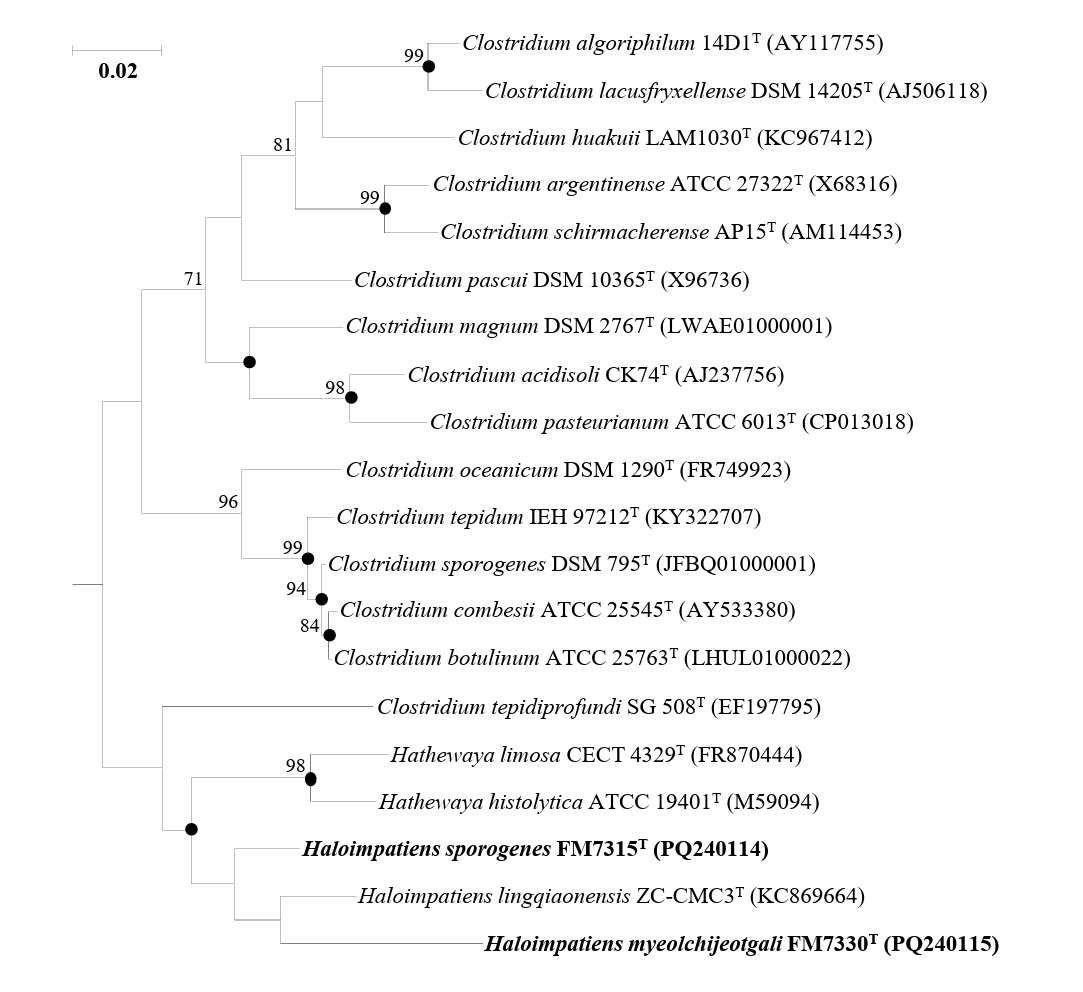
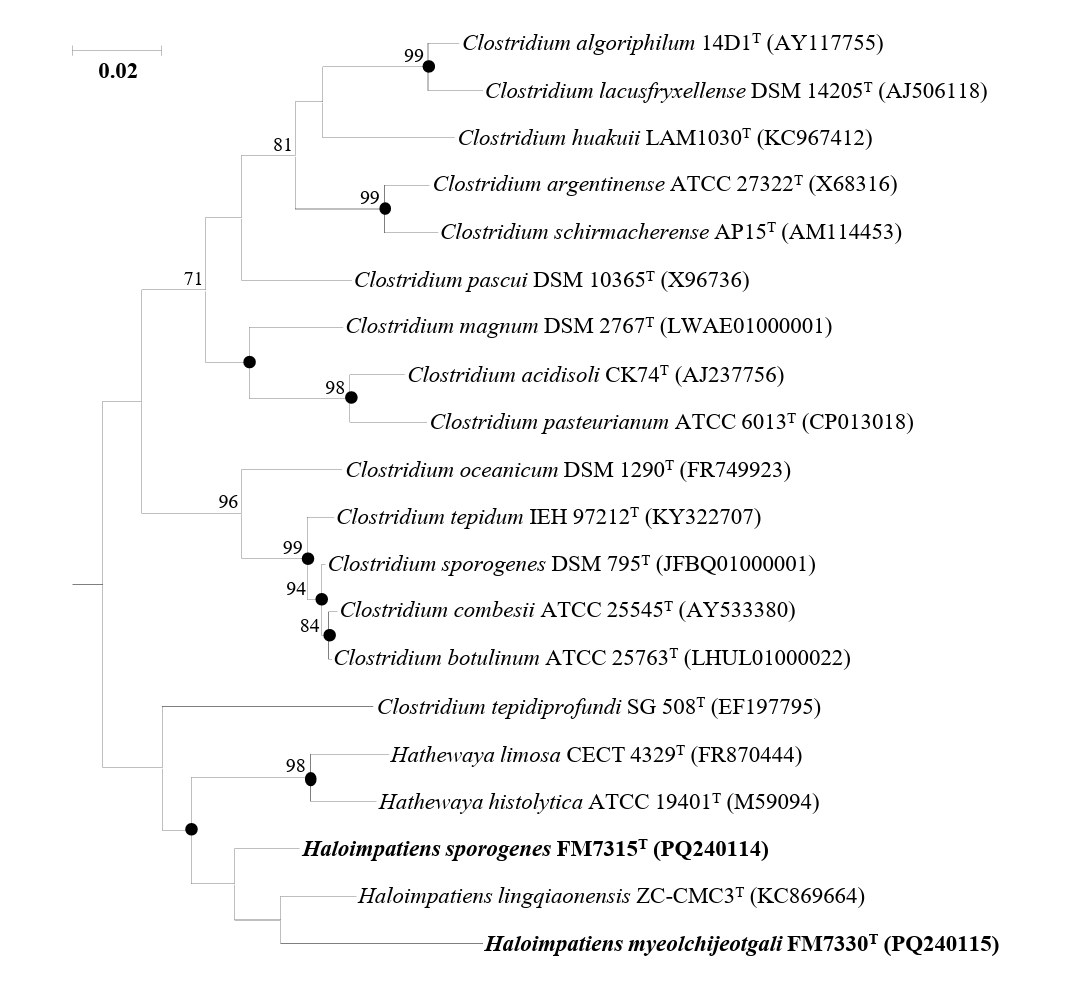
Analysis of the 16S rRNA gene sequence identities revealed that strains FM7315T and FM7330T were phylogenetically related to Haloimpatiens lingqiaonensis ZC-CMC3T, with 16S rRNA gene sequence similarities of 96.58% and 94.14%, respectively. The identity between strains FM7315T and FM7330T was 94.33%, all of which are significantly below the species threshold of 98.7% (Chun et al., 2018). Phylogenetic analysis based on the ML algorithm showed that strains FM7315T and FM7330T formed a distinct lineage, clustering with H. lingqiaonensis ZC-CMC3T but supported by a low bootstrap value (Fig. 1). Phylogenetic trees reconstructed using NJ and MP methods showed that FM7315T and FM7330T formed a distinct clade with H. lingqiaonensis ZC-CMC3T (Fig. S1).
Genome characteristics
The complete genomes of the strains FM7315T and FM7330T and the reference strain KCTC 15321T were obtained using genome sequencing and assembly (Fig. S2). The genome of strain FM7315T comprised one complete chromosome and one circular plasmid, whereas that of strain FM7330T included one complete chromosome and two circular plasmids. In contrast, the genome of strain KCTC 15321T contained only one complete chromosome. Genome quality assessment of strains FM7315T, FM7330T, and KCTC 15321T using CheckM (version 1.2.2) revealed completeness values of 99.36%, 98.31%, and 92.53%, respectively, highlighting the high quality and completeness of their genomes (Chklovski et al., 2023).
Genome-based phylogenomic analysis showed that strains FM7315T and FM7330T formed a distinct clade with H. lingqiaonensis KCTC 15321T (Fig. 2). To clarify their taxonomic positions at the genus level, AAI and POCP were analyzed (Table S1). Strains FM7315T and FM7330T showed high genomic identities with H. lingqiaonensis KCTC 15321T, which were above the genus delineation thresholds of 60–65% for AAI (Riesco and Trujillo, 2024) and 50% for POCP (Qin et al., 2014). These results supported the classification of FM7315T and FM7330T within the genus Haloimpatiens. To assess species-level relatedness, the ANI and dDDH values were calculated for strains FM7315T, FM7330T, and H. lingqiaonensis KCTC 15321T (Table S2). The ANI values between FM7315T and FM7330T and H. lingqiaonensis KCTC 15321T were 74.64% and 74.27%, respectively, which are below the species delineation threshold of 95–96% (Lee et al., 2016). Similarly, the dDDH values for the same pairs were 22.60% and 22.10%, respectively, both of which are below the 70% threshold for species delineation (Meier-Kolthoff et al., 2013). These results indicated that strains FM7315T and FM7330T represent novel species within the genus Haloimpatiens.
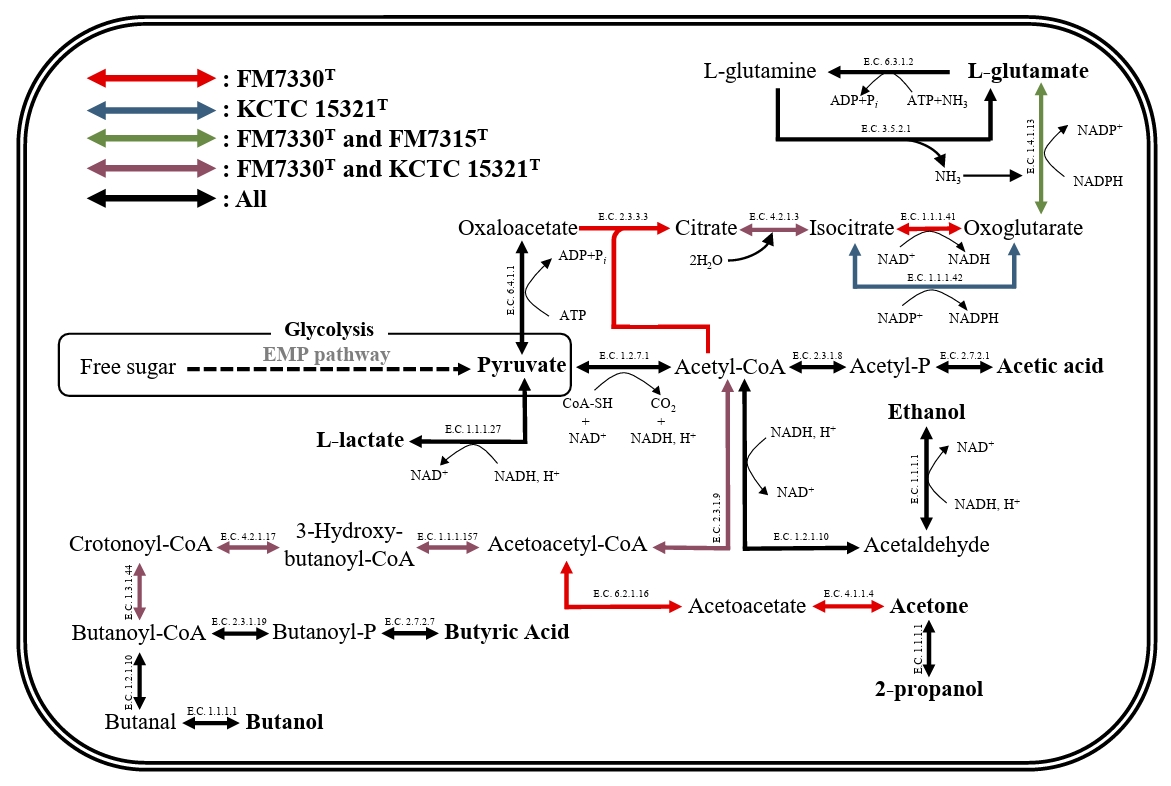
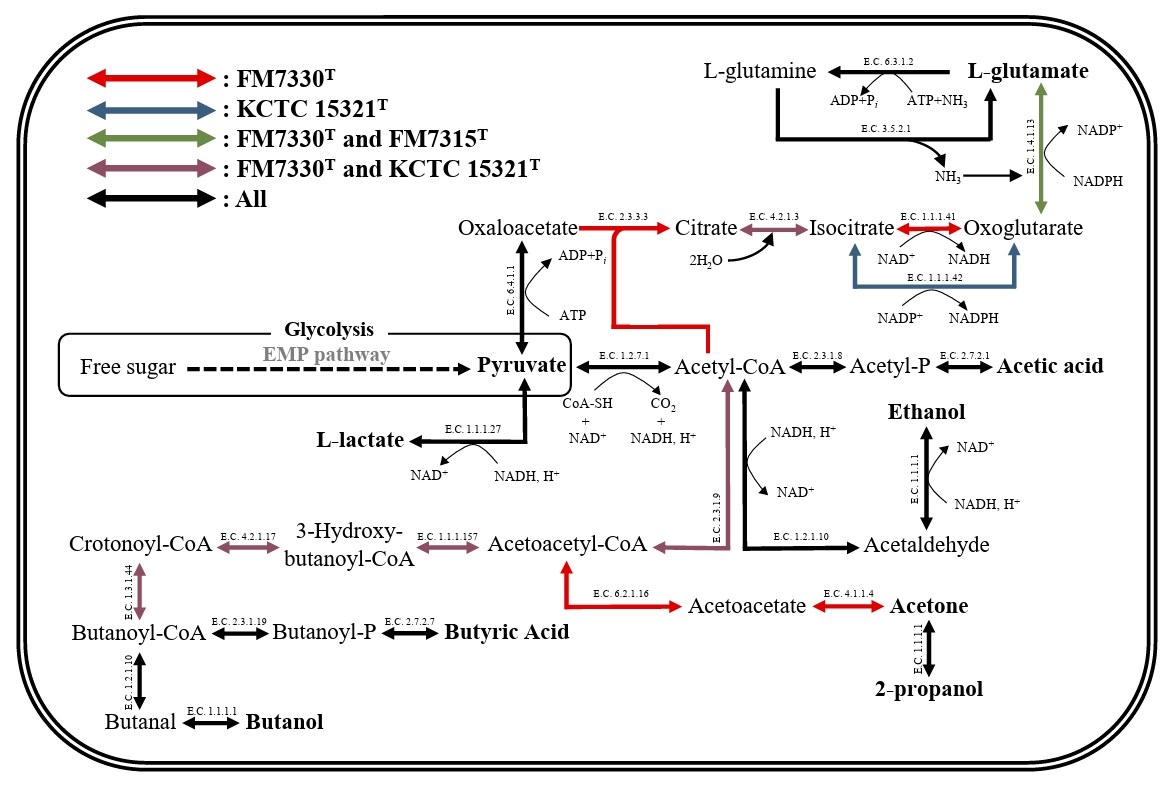
The genome sizes of strains FM7315T, FM7330T, and KCTC 15321T are 3.1, 4.2, and 3.3 Mbp, respectively, with G + C contents of 29.5%, 28.0%, and 31.0%, respectively. These values are consistent with the genome sizes and G + C contents of species within the genus Haloimpatiens currently registered in the GenBank database (3.3–4.2 Mbp and 30–31%). The genomes of strains FM7315T, FM7330T, and KCTC 15321T harbored 2,893, 3,769, and 3,182 genes, respectively, including 2,728, 3,581, and 2,936 protein-coding genes; 93, 94, and 91 tRNA genes; and 30, 31, and 27 rRNA genes, respectively. The genomes of strains FM7315T, FM7330T, and KCTC 15321T were predicted to contain 43, 71, and 43 genes, respectively, encoding various CAZy. Strain FM7330T had a higher ability to degrade polysaccharides than strains FM7315T and KCTC 15321T. Especially, strain FM7330T uniquely possesses genes involved in 6 glycoside hydrolase families (EC 3.2.1.-), two glycosyltransferase families (EC 2.4.1.-), two auxiliary activity families (EC 1.1.3.- and EC 1.11.1.-), and two carbohydrate-binding module families, which contribute to their ability to degrade diverse carbohydrates and support their metabolic functions during food fermentation. KEGG analysis revealed that strains FM7315T, FM7330T, and KCTC 15321T contained genes associated with lactic acid and ethanol production, suggesting their potential role in fermentation-related metabolic processes under anaerobic conditions (Fig. 3). In addition, metabolic genes related to butyric acid production were identified in the genomes of strains FM7330T and KCTC 15321T, while strain FM7315T was found to possess only a subset of the associated genes. Nevertheless, butyric acid was detected in all three strains, suggesting that strain FM7315T may harbor alternative genes or pathways involved in its biosynthesis. Strains FM7315T and FM7330T contained a gene encoding glutamate dehydrogenase, an enzyme important for glutamate production. Glutamate is a key compound that gives food an umami flavor. This suggests that strains FM7315T and FM7330T may enhance umami taste during food fermentation. Moreover, KEGG pathway analysis identified genes related to flagellar assembly and motility in the genomes of FM7315T and FM7330T (Table S3).
Physiological and morphological characteristics
Strains FM7315T and FM7330T grew well on BS agar, BHI agar, and YPD agar but did not grow on R2A agar. The growth conditions for strain FM7315T were exhibited within a temperature range of 20–37°C (optimal 30°C), NaCl concentration range of 0–2% (optimal 0%), and pH range of 5.0–8.0 (optimal 6.5). In contrast, strain FM7330T exhibited slightly different growth conditions, with a temperature range of 25–45°C (optimal 40°C), NaCl concentration range of 0–4% (optimal 2%), and pH range of 5.0–9.0 (optimal 7.0). Strains FM7315T and FM7330T are Gram-positive and spore forming. Cells observed under an electron microscope were rod-shaped, measuring 1.7–5.7 µm in length and 0.4–0.9 µm in width for strain FM7315T, and 2.1–7.4 µm in length and 0.7–2.1 µm in width for strain FM7330T. Motility tests on semi-solid BL agar confirmed that both FM7315T and FM7330T were motile. Aerobic growth was not observed after 21 d of incubation under optimal conditions.
Strains FM7315T and FM7330T shared several phenotypic traits with Haloimpatiens lingqiaonensis KCTC 15321T, such as catalase and oxidase activity and the inability to hydrolyze Tween 20 and Tween 80. However, the three strains exhibited distinct characteristics (Table 3). Strains FM7315T and FM7330T hydrolyzed starch but not casein, whereas strain KCTC 15321T hydrolyzed casein but not starch. API ZYM testing revealed that all three strains were positive for acid phosphatase and naphthol-AS-BI phosphohydrolase. Strain FM7330T exhibited esterase (C4) and esterase lipase (C8) activity. Strain FM7315T utilizes D-fructose, D-glucose, and D-maltose, whereas strain FM7330T metabolizes D-ribose and pyruvate. These results were further supported by the identification of relevant metabolic genes in the genomes of both strains, particularly by the observation that strain FM7330T harbored a greater number of CAZy families, consistent with its broader substrate utilization profile. Butyrate was uniquely produced by strains FM7315T and FM7330T, whereas it was not detected in strain ZC-CMC3T. Additionally, strains FM7315T and ZC-CMC3T produced ethanol, whereas strain FM7330T did not.
Chemotaxonomic characteristics
The polar lipid and cellular fatty acid profiles of strains FM7315T and FM7330T differed from those of Haloimpatiens lingqiaonensis KCTC 15321T. Cellular fatty acid analysis revealed that C16:0 was abundant in all three strains (Table S4). In contrast, C14:0 was detected only in FM7315T and KCTC 15321T. FM7315T and FM7330T shared C18:1 ω9c as a major component, whereas this fatty acid was not prominent in KCTC 15321T. Additionally, FM7315T possessed Summed Feature 3 (C16:1 ω7c/C16:1 ω6c) and Summed Feature 8 (C18:1 ω7c/C18:1 ω6c) as major components, whereas FM7330T contained C17:1 ω8c. Strains FM7315T and FM7330T both harbor two major polar lipids, DPG and PG. In addition, strains FM7315T and FM7330T shared five unidentified polar lipids (ULs) and two aminophospholipids (APLs) not observed in strain ZC-CMC3T. Strain FM7315T also possessed an additional GL, whereas FM7330T contained two additional APLs. These compositional differences are common within the family Clostridiaceae (Chaikitkaew et al., 2022; Flaiz et al., 2020; Xu et al., 2019), and are presumed to be influenced by environmental factors related to the isolation of strains. These variations are likely attributable to the environmental factors associated with the respective sources of isolation.
Taxonomic Conclusion
Description of Haloimpatiens sporogenes sp. nov.
Haloimpatiens sporogenes (spo.ro’ge.nes. Gr. fem. n. spora, a spore; Gr. v. gennao, produce; and N.L. masc. part. adj. sporogenes, spore-producing). Cells are strictly anaerobic, spore forming, Gram-positive, motile, rod-shaped, 1.7–5.7 µm in length, and 0.4–0.9 µm in width. Growth is observed on BS agar at temperatures of 20–37°C (optimum 30°C), NaCl concentrations of 0–2% (optimum 0%), and with a pH range of 5.0–8.0 (optimum 6.5). Cells hydrolyze starch; assimilate D-fructose, D-glucose, and D-maltose as sole carbon sources; and exhibit positive enzymatic activity for acid phosphatase and naphthol-AS-B1 phosphohydrolase. Acetic, butyric, lactic acids, and ethanol were the major fermentation products. The major fatty acids are C14:0, C16:0, C18:1 ω9c, Summed Feature 3 (C16:1 ω7c/C16:1 ω6c), and Summed Feature 8 (C18:1 ω7c/C18:1 ω6c), while the major polar lipids are diphosphatidylglycerol, phosphatidylglycerol, glycolipid, aminophospholipids and unidentified lipids. The type strain is FM7315T (= KCTC 25939T = JCM 37574T), isolated from Myeolchi-jeot, a traditional Korean fermented anchovy. The total genome size, including both the chromosome and the plasmid, is 3,052,517 bp, with a G + C content of 29.7 mol%. The GenBank accession numbers for the 16S rRNA gene and complete genome sequences are PQ240114 and CP175761, respectively.
Description of Haloimpatiens myeolchijeotgali sp. nov.
Haloimpatiens myeolchijeotgali (mye.ol.chi.je.ot'ga.li. N.L. gen. n. myeolchijeotgali, Myeolchi-jeotgal, traditional Korean fermented anchovy). Cells are strictly anaerobic, spore forming, Gram-positive, motile, rod-shaped, 2.1–6.3 µm in length and 0.7–2.1 µm in width. Growth is observed on BS agar at temperatures of 25–45°C (optimum 40°C), NaCl concentration of 0–4% (optimum 2%), and with a pH range of 5.0–9.0 (optimum 7.0). Cells hydrolyze starch; assimilate pyruvate, D-fructose, D-glucose, D-maltose, and D-ribose as sole carbon sources; and exhibit positive enzymatic activity for acid phosphatase, naphthol-AS-B1 phosphohydrolase, esterase, and esterase lipase. Acetic, butyric, and lactic acids are the major fermentation products. The major fatty acids are C16:0, C17:1 ω8c, and C18:1 ω9c, while the major polar lipids are diphosphatidylglycerol, phosphatidylglycerol, aminophospholipids, and unidentified lipids. The type strain is FM7330T (= KCTC 25938T = JCM 37575T), isolated from Myeolchi-jeot, a traditional Korean fermented anchovy. The total genome size, comprising one chromosome and two plasmids, is 4,194,114 bp, with a G + C content of 28.0 mol%. The GenBank accession numbers for the 16S rRNA gene and complete genome sequences are PQ240115 and JBHWAB000000000, respectively.
Acknowledgments
This work was supported by the Pukyong National University Industry-university Cooperation Research Fund in 2023 (202312360001).
Conflict of Interest
The authors have no conflicts of interest.
Supplementary Information
The online version contains supplementary material available at https://doi.org/10.71150/jm.2504009.
Table S1.
Comparison of the genomic identities of strains FM7315T and FM7330T with those of their phylogenetically closely related species in the genus Haloimpatiens and the type species of the genus Clostridium, using AAI and POCP analysis. Strains: 1, Haloimpatiens sporogenes FM7315T; 2, Haloimpatiens myeolchijeotgali FM7330T; 3, Haloimpatiens lingqiaonensis KCTC 15321T; 4, Clostridium butyricum DSM 10702T
jm-2504009-Supplementary-Table-S1.pdf
Table S2.
Pairwise comparison of average nucleotide identity (ANI) and digital DNA-DNA hybridization (dDDH) values between strains FM7315T and FM7330T and the type strain of phylogenetically closely related species in the genus Haloimpatiens. Strains: 1, Haloimpatiens sporogenes FM7315T; 2, Haloimpatiens myeolchijeotgali FM7330T; 3, Haloimpatiens lingqiaonensis KCTC 15321T
jm-2504009-Supplementary-Table-S2.pdf
Table S4.
Comparison of cellular fatty acid compositions (%) between the two novel strains and Haloimpatiens lingqiaonensis KCTC 15321T. Strains: 1, H. sporogenes FM7315T, 2, FM7330T, 3, H. lingqiaonensis KCTC 15321T. The majority of fatty acid values (> 5%) are highlighted in bold. tr, trace element (< 1%); –, not detected
jm-2504009-Supplementary-Table-S4.pdf
Fig. S1.
Neighbor-joining (A) and maximum-parsimony (B) trees based on 16S rRNA gene sequences showing the phylogenetic relationships of strains H. sporogenes FM7315T, H. myeochijeotgali FM7330T, and their closely related taxa. Bootstrap values with more than 70% are shown on the nodes as percentages of 1,000 replicates. Bacillus subtilis NCIB 3610T (ABQL01000001) was selected as the outgroup.
jm-2504009-Supplementary-Fig-S1.pdf
Fig. S4.
Two-dimensional thin-layer chromatoGrams showing the compositions of total polar lipids (A), glycolipids (B), aminolipids (C), and phospholipids (D) in strains FM7315T. Total lipids were stained with 90% ethanolic molybdophosphoric acid, glycolipids with α-naphthol and sulfuric acid, amino lipids with ninhydrin, and phospholipids with both ninhydrin and the Dittmer-Lester reagent. Solvent system: (I) chloroform-methanol-water (65:25:4, v/v/v); (II) chloroform-acetic acid-methanol-water (80:15:12:4, v/v/v/v). DPG, diphosphatidylglycerol; PG, phosphatidylglycerol; APL, aminophospholipid; GL, unidentified glycolipid; UL, unidentified polar lipid.
jm-2504009-Supplementary-Fig-S4.pdf
Fig. S5.
Two-dimensional thin-layer chromatoGrams showing the compositions of total polar lipids (A), glycolipids (B), aminolipids (C), and phospholipids (D) in strains FM7330T. Total lipids were stained with 90% ethanolic molybdophosphoric acid, glycolipids with α-naphthol and sulfuric acid, amino lipids with ninhydrin, and phospholipids with both ninhydrin and the Dittmer-Lester reagent. Solvent system: (I) chloroform-methanol-water (65:25:4, v/v/v); (II) chloroform-acetic acid-methanol-water (80:15:12:4, v/v/v/v). DPG, diphosphatidylglycerol; PG, phosphatidylglycerol; APL, aminophospholipid; UL, unidentified polar lipid.
jm-2504009-Supplementary-Fig-S5.pdf
Fig. 1.A maximum-likelihood tree based on 16S rRNA gene sequences showing the phylogenetic relationships of strains H. sporogenes FM7315T, H. myeolchijeotgali FM7330T, and their closely related taxa. Bootstrap values with more than 70% are shown on the nodes as percentages of 1,000 replicates. Filled circles (●) indicate nodes that were also identified in trees reconstructed with the neighbor-joining and maximum-likelihood algorithms. Bacillus subtilis NCIB 3610T (ABQL01000001) was selected as the outgroup. The scale bar, 50 changes per nucleotide position.
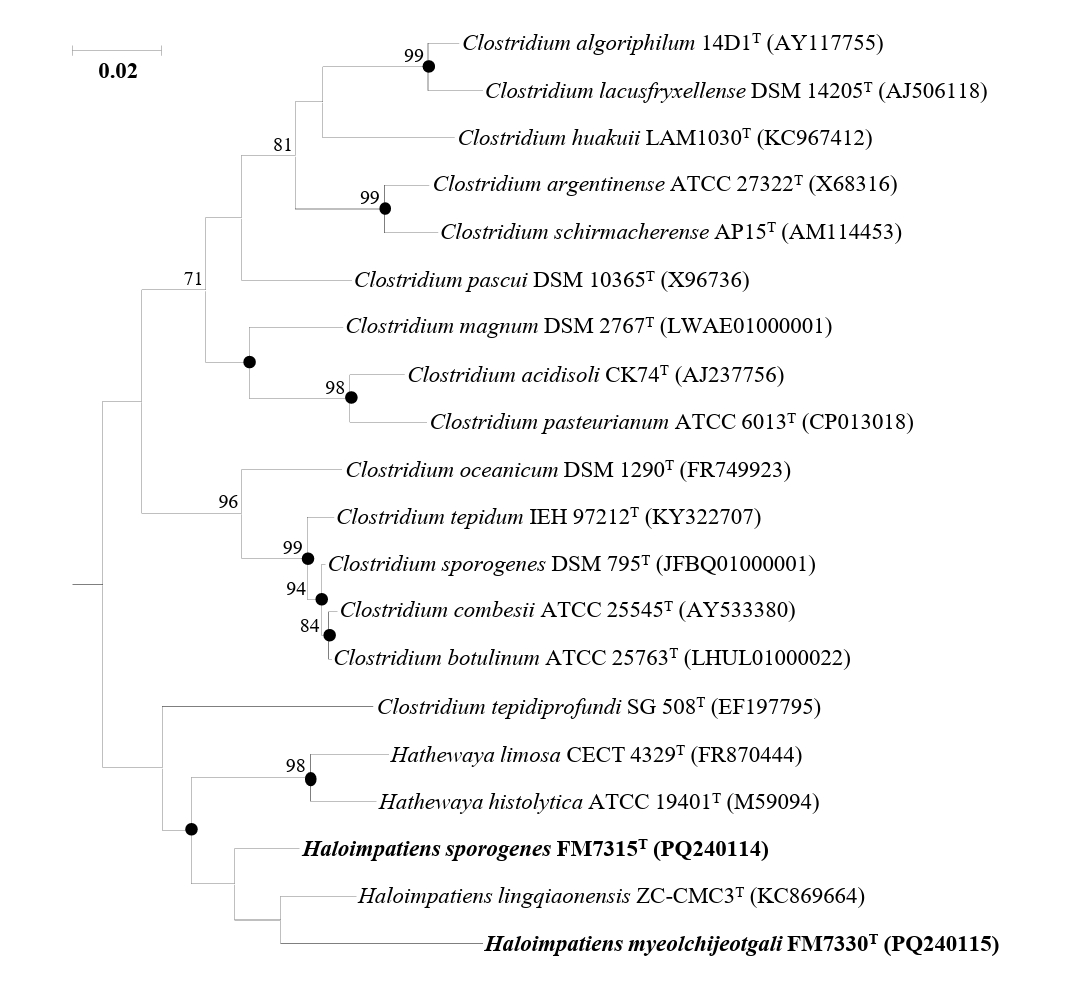
Fig. 2.A maximum-likelihood tree showing the phylogenomic relationships of strains H. sporogenes FM7315T (CP175761), H. myeolchijeotgali FM7330T (JBHWAB000000000), and related taxa, based on concatenated 92 core gene sequences. Bootstrap values with more than 70% are shown on the nodes as percentages of 1,000 replicates. Bacillus subtilis DSM 10T (JAEPVU000000000) was selected as the outgroup. The scale bar, 0.1 changes per nucleotide position.

Fig. 3.Carbon metabolic pathways of the strains FM7315T, FM7330T, and KCTC 15321T for fermentation. The arrow colors indicate the metabolic pathways as follows: red represents pathways exclusive to FM7330T, blue represents those unique to KCTC 15321T, green represents pathways shared between FM7330T and FM7315T, purple represents those common to FM7330T and KCTC 15321T, and black represents pathways present in all three strains.
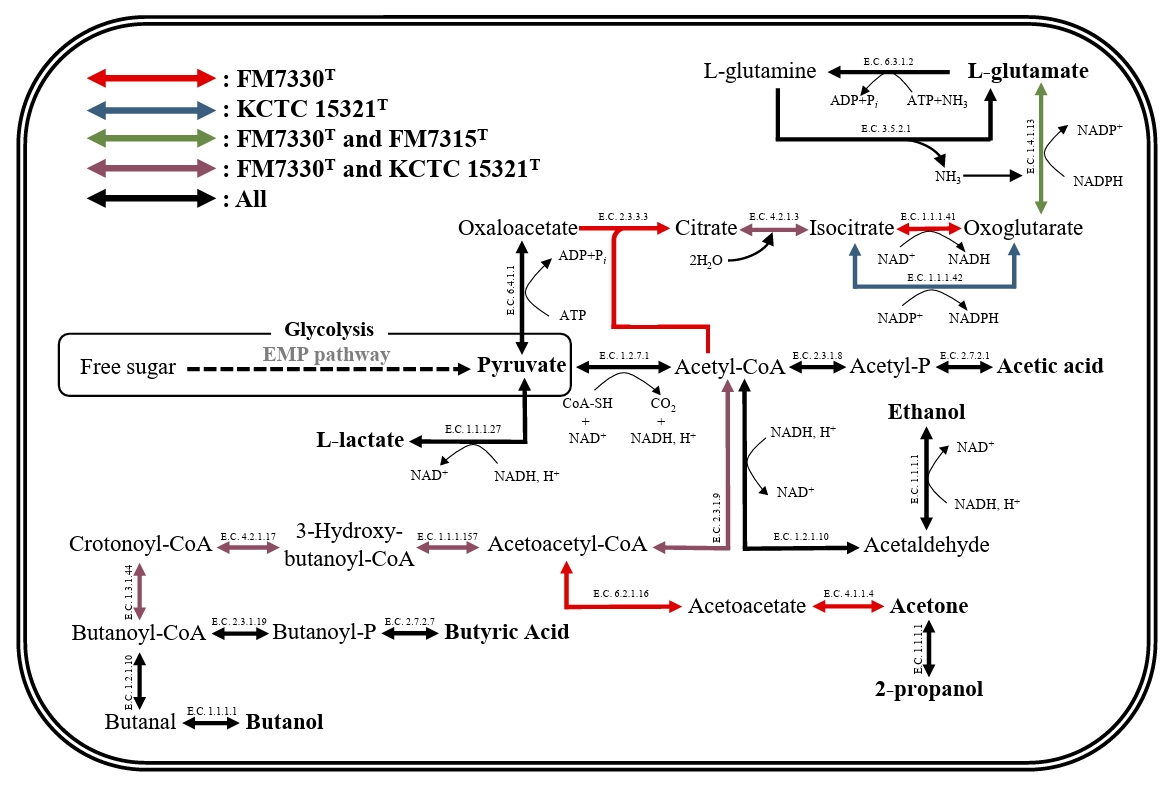
Table 1.Genomic features of the two novel strains and Haloimpatiens lingqiaonensis KCTC 15321T. Strains; 1, H. sporogenes FM7315T (this study); 2, H. myeolchijeotgali FM7330T (this study); 3, H. lingqiaonensis KCTC 15321T (this study)
|
Feature |
1 |
2 |
3 |
|
Genome size (Mb) |
3.0 |
4.2 |
3.5 |
|
Status |
Complete |
Complete |
Complete |
|
No. of contigs |
2 |
3 |
1 |
|
Chromosome |
1 |
1 |
1 |
|
Plasmid |
1 |
2 |
0 |
|
G + C contents (mol%) |
29.5 |
28.0 |
31.0 |
|
N50 |
3.0 Mb |
4.0 Mb |
3.5 Mb |
|
No. of total genes |
2,893 |
3,769 |
3,182 |
|
No. of protein-coding genes |
2,728 |
3,581 |
2,936 |
|
No. of total RNA genes |
127 |
129 |
123 |
|
No. of tRNA genes |
93 |
94 |
91 |
|
No. of rRNA genes |
30 |
31 |
27 |
|
No. of total CAZy genes |
43 |
71 |
43 |
|
Glycoside hydrolase |
12 |
22 |
10 |
|
Glycosyltransferase |
20 |
29 |
23 |
|
Carbohydrate esterase |
7 |
10 |
6 |
|
Auxiliary activity |
1 |
8 |
0 |
|
Carbohydrate-binding module |
3 |
2 |
4 |
|
No. of ncRNA genes |
4 |
4 |
5 |
|
No. of pseudogenes |
38 |
59 |
123 |
|
Coverage (×) |
175 |
114 |
65 |
|
GenBank accession NO. |
CP175761 |
JBHWAB000000000 |
CP172402 |
Table 2.Comparison of phenotypic characteristics between the two novel strains and Haloimpatiens lingqiaonensis KCTC 15321T. Strains; 1, H. sporogenes FM7315T (this study); 2, H. myeolchijeotgali FM7330T (this study); 3, H. lingqiaonensis KCTC 15321T (this study). All strains were positive for the following: enzyme activities of acid phosphatase and naphthol-AS-B1 phosphohyderolase. All strains were negative for the following: hydrolysis of Tween 20 and Tween 80; enzyme activities of leucine arylamidase, α-chymotrypsin, lipase, valine arylamidase, cystine arylamidase, trypsin, α-galactosidase, β-galactosidase, β-glucuronidase, α-glucosidase, β-glucosidase, N-acetyl-β-glucosaminidase, α-mannosidase, catalase and oxidase; and utilization of L-arabinose, D-galactose, D-mannitol, D-xylose, L-xylose, formic acid, acetic acid, butyric acid, propionic acid, fumaric acid, lactic acid, malic acid, succinic acid, ethanol, butanol, propanol, arginine, asparagine, aspartate, glutamine, glycine, tryptophan, tyrosine, alanine, histidine, isoleucine, lysine, methionine, phenylalanine, serine, threonine, and valine as single carbon sources. +, Positive; –, negative. A, acetate; B, butyrate; E, ethanol; F, formate; L, lactate
|
Characteristics |
1 |
2 |
3 |
|
Isolation Source†
|
Myeolchi-jeot |
Myeolchi-jeot |
Paper-mill wastewater |
|
Growth range† of: |
|
|
|
|
Temperature (optimum, ℃) |
20–37 (30) |
25–45 (40) |
25–48 (43) |
|
pH (optimum) |
5.0–8.0 (6.5) |
5.0–9.0 (7) |
5.5–8.0 (7) |
|
NaCl (optimum, %) |
0–2 (0) |
0–4 (2) |
0–3 (0) |
|
Spore forming†
|
+ |
+ |
– |
|
Hydrolysis† of: |
|
|
|
|
Starch |
+ |
+ |
– |
|
Casein |
– |
– |
+ |
|
Enzyme activities (API ZYM): |
|
|
|
|
Alkaline phosphate |
– |
– |
+ |
|
Esterase (C4) |
– |
+ |
– |
|
Esterase lipase (C8) |
– |
+ |
– |
|
α-Fucosidase |
|
– |
+ |
|
Utilization† of: |
|
|
|
|
Pyruvate |
- |
+ |
– |
|
D-Fructose |
+ |
+ |
– |
|
D-Glucose |
+ |
+ |
– |
|
D-Maltose |
+ |
+ |
– |
|
D-Ribose |
– |
+ |
– |
|
Fermentation products†*
|
A, B, E, L |
A, B, L |
F, A, E, L |
References
- Agilent Technologies Inc. 2016. Single Quad LC/MS analysis of organic acids using an Agilent HiPlex column. Agilent Technologies Inc.
- Anani H, Alou MT, Fontanini A, Raoult D, Lagier JC, et al. 2020. Taxono-genomics and description of Haloimpatiens massiliensis sp. nov., a new bacterium isolated from the gut of a healthy infant. New Microbes New Infect. 33: 100631.ArticlePubMedPMC
- Chaikitkaew S, In-Chan S, Singkhala A, Tukanghan W, Mamimin C, et al. 2022. Clostridium thailandense sp. nov., a novel CO₂-reducing acetogenic bacterium isolated from peatland soil. Int J Syst Evol Microbiol. 72: 005254.Article
- Chklovski A, Parks DH, Woodcroft BJ, Tyson GW. 2023. CheckM2: a rapid, scalable and accurate tool for assessing microbial genome quality using machine learning. Nat Methods. 20: 1203–1212. ArticlePubMedPDF
- Chun J, Oren A, Ventosa A, Christensen H, Arahal DR, et al. 2018. Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int J Syst Evol Microbiol. 68: 461–466. ArticlePubMed
- Felsenstein J. 1981. Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol. 17: 368–376. ArticlePubMedPDF
- Fitch WM. 1971. Toward defining the course of evolution: minimum change for a specific tree topology. Syst Biol. 20: 406–416. Article
- Flaiz M, Baur T, Brahner S, Poehlein A, Daniel R, et al. 2020. Caproicibacter fermentans gen. nov., sp. nov., a new caproate-producing bacterium and emended description of the genus Caproiciproducens. Int J Syst Evol Microbiol. 70: 4269–4279. ArticlePubMed
- Gerhardt P, Murray RGE, Wood WA, Krieg NR. 1994. Methods for General and Molecular Bacteriology. American Society for Microbiology.
- Gomori G. 1995. Preparation of buffers for use in enzyme studies. In Colowick SP, Kaplan NO. (eds.), Methods in Enzymology, pp. 138–146. Academic Press.
- Gram C. 1884. Ueber die isolirte Färbung der Schizomyceten in Schnitt- und Trockenpräparaten. Fortschr Med. 2: 185–189.
- Grant JR, Enns E, Marinier E, Mandal A, Herman EK, et al. 2023. Proksee: in-depth characterization and visualization of bacterial genomes. Nucleic Acids Res. 51: W484–W492. ArticlePubMedPMCPDF
- Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 41: 95–98.
- Hernández-Eugenio G, Fardeau ML, Cayol JL, Patel BK, Thomas P, et al. 2002. Clostridium thiosulfatireducens sp. nov., a proteolytic, thiosulfate- and sulfur-reducing bacterium isolated from an upflow anaerobic sludge blanket (UASB) reactor. Int J Syst Evol Microbiol. 52: 1461–1468. ArticlePubMed
- Kanehisa M, Goto S. 2000. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28: 27–30. ArticlePubMedPMC
- Kim J, Na SI, Kim D, Chun J. 2021. UBCG2: Up-to-date bacterial core genes and pipeline for phylogenomic analysis. J Microbiol. 59: 609–615. ArticlePubMedPDF
- Kolmogorov M, Yuan J, Lin Y, Pevzner PA. 2019. Assembly of long, error-prone reads using repeat graphs. Nat Biotechnol. 37: 540–546. ArticlePubMedPDF
- Lane DJ. 1991. 16S/23S rRNA sequencing. In Stackebrandt E, Goodfellow M. (eds.), Nucleic acid techniques in bacterial systematics, pp. 115–175. John Wiley and Sons.
- Lee I, Kim YO, Park SC, Chun J. 2016. OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int J Syst Evol Microbiol. 66: 1100–1103. ArticlePubMed
- Lee KE, Lee SM, Choi YH, Hurh BS, Kim YS. 2013. Comparative volatile profiles in soy sauce according to inoculated microorganisms. Biosci Biotechnol Biochem. 77: 2192–2200. ArticlePubMed
- Meier-Kolthoff JP, Auch AF, Klenk HP, Göker M. 2013. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics. 14: 60.ArticlePubMedPMCPDF
- Minnikin DE, Patel PV, Alshamaony L, Goodfellow M. 1977. Polar lipid composition in the classification of Nocardia and related bacteria. Int J Syst Evol Microbiol. 27: 104–117. Article
- Parte AC, Sardà Carbasse J, Meier-Kolthoff JP, Reimer LC, Göker M. 2020. List of Prokaryotic names with Standing in Nomenclature (LPSN) moves to the DSMZ. Int J Syst Evol Microbiol. 70: 5607–5612. ArticlePubMedPMC
- Pruesse E, Peplies J, Glöckner FO. 2012. SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics. 28: 1823–1829. ArticlePubMedPMCPDF
- Qin QL, Xie BB, Zhang XY, Chen XL, Zhou BC, et al. 2014. A proposed genus boundary for the prokaryotes based on genomic insights. J Bacteriol. 196: 2210–2215. ArticlePubMedPMCLink
- Reynolds J, Moyes R, Breakwell DP. 2009. Differential staining of bacteria: endospore stain. Curr Protoc Microbiol. 15: A-3J.ArticleLink
- Riesco R, Trujillo ME. 2024. Update on the proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int J Syst Evol Microbiol. 74: 006300.ArticlePubMedPMC
- Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 4: 406–425. ArticlePubMed
- Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular Cloning: A Laboratory Manual, 2nd edn. Cold Spring Harbor Laboratory Press.
- Sasser M. 1990. Identification of bacteria by gas chromatography of cellular fatty acids. MIDI Technical Note 101, MIDI Inc.
- Tamura K, Stecher G, Kumar S. 2021. MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol. 38: 3022–3027. ArticlePubMedPMCPDF
- Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Nawrocki EP, et al. 2016. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 44: 6614–6624. ArticlePubMedPMC
- Tittsler RP, Sandholzer LA. 1936. The use of semi-solid agar for the detection of bacterial motility. J Bacteriol. 31: 575–580. ArticlePubMedPMCLink
- Wiegel J, Tanner RA, Rainey FA. 2006. An introduction to the family Clostridiaceae. The Prokaryotes. 4: 654–678. Article
- Wu D, Zhang NF, Sun C, Zhang WW, Han SB, et al. 2016. Haloimpatiens lingqiaonensis gen. nov., sp. nov., an anaerobic bacterium isolated from paper-mill wastewater. Int J Syst Evol Microbiol. 66: 628–632. ArticlePubMed
- Xu PX, Chai LJ, Qiu T, Zhang XJ, Lu ZM, et al. 2019. Clostridium fermenticellae sp. nov., isolated from the mud in a fermentation cellar for the production of the Chinese liquor, baijiu. Int J Syst Evol Microbiol. 69: 859–865. ArticlePubMed
- Yang F, Wang H, Chen LQ, Zhou N, Lu JJ, et al. 2024. Clostridium lapidicellarium sp. nov. and Clostridium moutaii sp. nov., two species isolated from fermentation cellar-producing sauce-flavour Chinese baijiu. Int J Syst Evol Microbiol. 74: 006580.ArticlePubMedPMC
Citations
Citations to this article as recorded by





 ePub Link
ePub Link Cite this Article
Cite this Article

 MSK
MSK