Articles
- Page Path
- HOME > J. Microbiol > Volume 63(11); 2025 > Article
-
Review
Metabolite-mediated mechanisms linking the urinary microbiome to bladder cancer - Thu Anh Trần, Ho Young Lee, Hae Woong Choi*
-
Journal of Microbiology 2025;63(11):e2509001.
DOI: https://doi.org/10.71150/jm.2509001
Published online: November 30, 2025
Division of Life Sciences, Korea University, Seoul 02841, Republic of Korea
- *Correspondence Hae Woong Choi haewoongchoi@korea.ac.kr
© The Microbiological Society of Korea
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/4.0) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
- ABSTRACT
- Introduction
- Urobiomes in Healthy Subjects
- Methodological Influences on Bladder Cancer Urobiome Studies
- Urobiome Diversity in Bladder Cancer Patients
- Microbial Metabolites and Bladder Cancer Progression
- The Role of the Microbiota–Metabolite–Immunity Axis in Bladder Cancer and Precision Therapeutic Strategies
- Conclusion and Future Perspectives
- Notes
- References
ABSTRACT
- Bladder cancer is the most common malignancy of the urinary tract and is a major health burden globally. Recent advances in microbiome research have revealed that the urinary tract harbors a resident microbial community, overturning the long-held belief in its sterility. Increasing evidence suggests that microbial dysbiosis and microbially derived metabolites contribute to bladder cancer carcinogenesis, progression, and therapeutic responses. Distinct microbial signatures have been observed in bladder cancer patients, with notable differences across disease stages and between primary and recurrent cases. Mechanistic studies have demonstrated that microbe-associated metabolites and toxins can drive DNA damage, chronic inflammation, extracellular matrix remodeling, and epithelial–mesenchymal transition. In addition, biofilm formation allows bacteria to evade immune responses and promotes persistent inflammation, creating a tumor-permissive niche. Beyond pathogenesis, microbial activity also influences therapeutic outcomes; for instance, some microbial pathways can inactivate frontline chemotherapy, while others generate metabolites with anti-tumor properties. Collectively, these patterns define a microbiota–metabolite–immunity axis, presenting opportunities for precision oncology. Targeting microbial pathways, profiling urinary microbiota, and harnessing beneficial metabolites offer promising advancements in biomarker discovery, prognostic refinement, and the development of novel therapeutic strategies for bladder cancer.
Introduction
Urobiomes in Healthy Subjects
Methodological Influences on Bladder Cancer Urobiome Studies
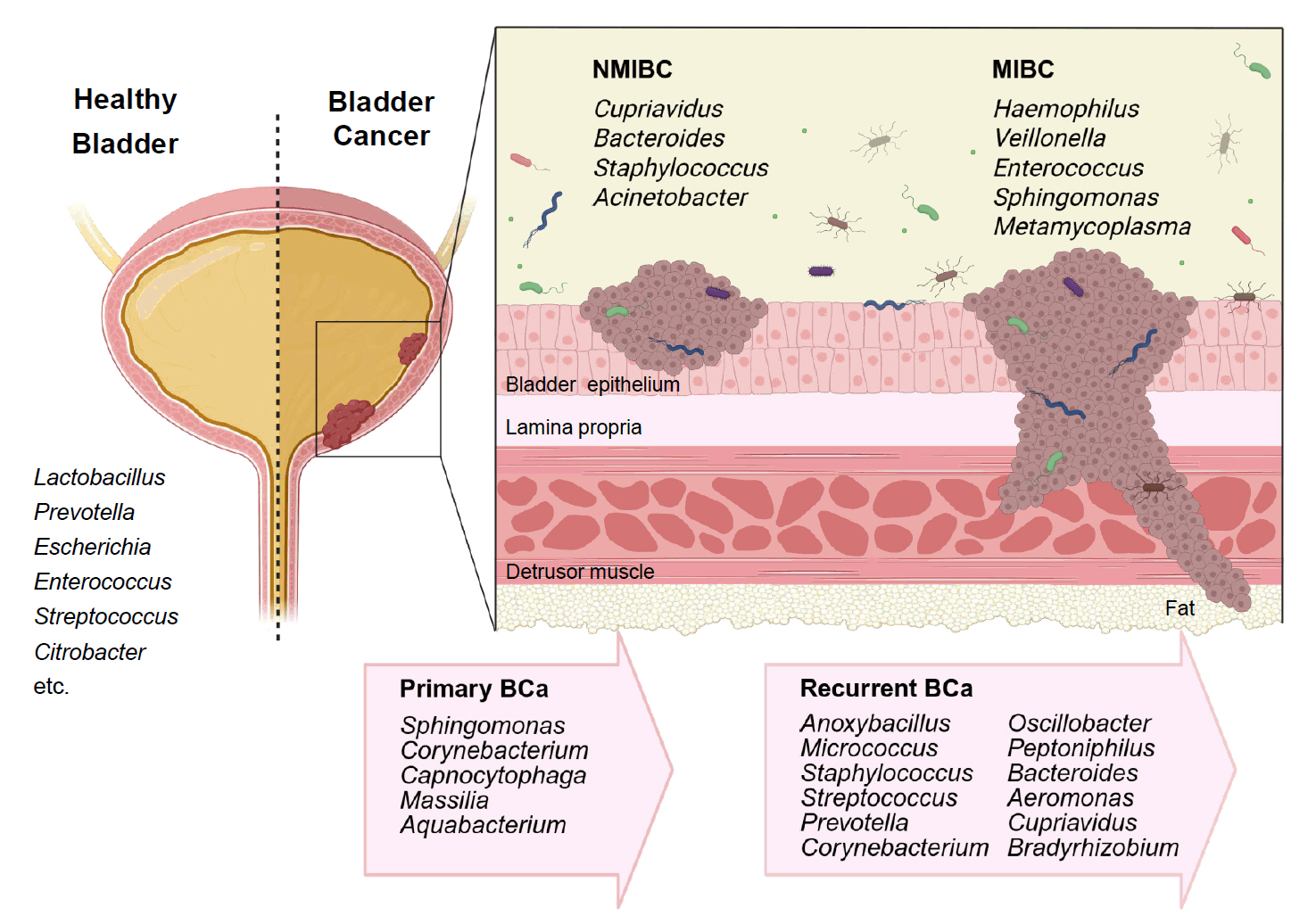
Urobiome Diversity in Bladder Cancer Patients
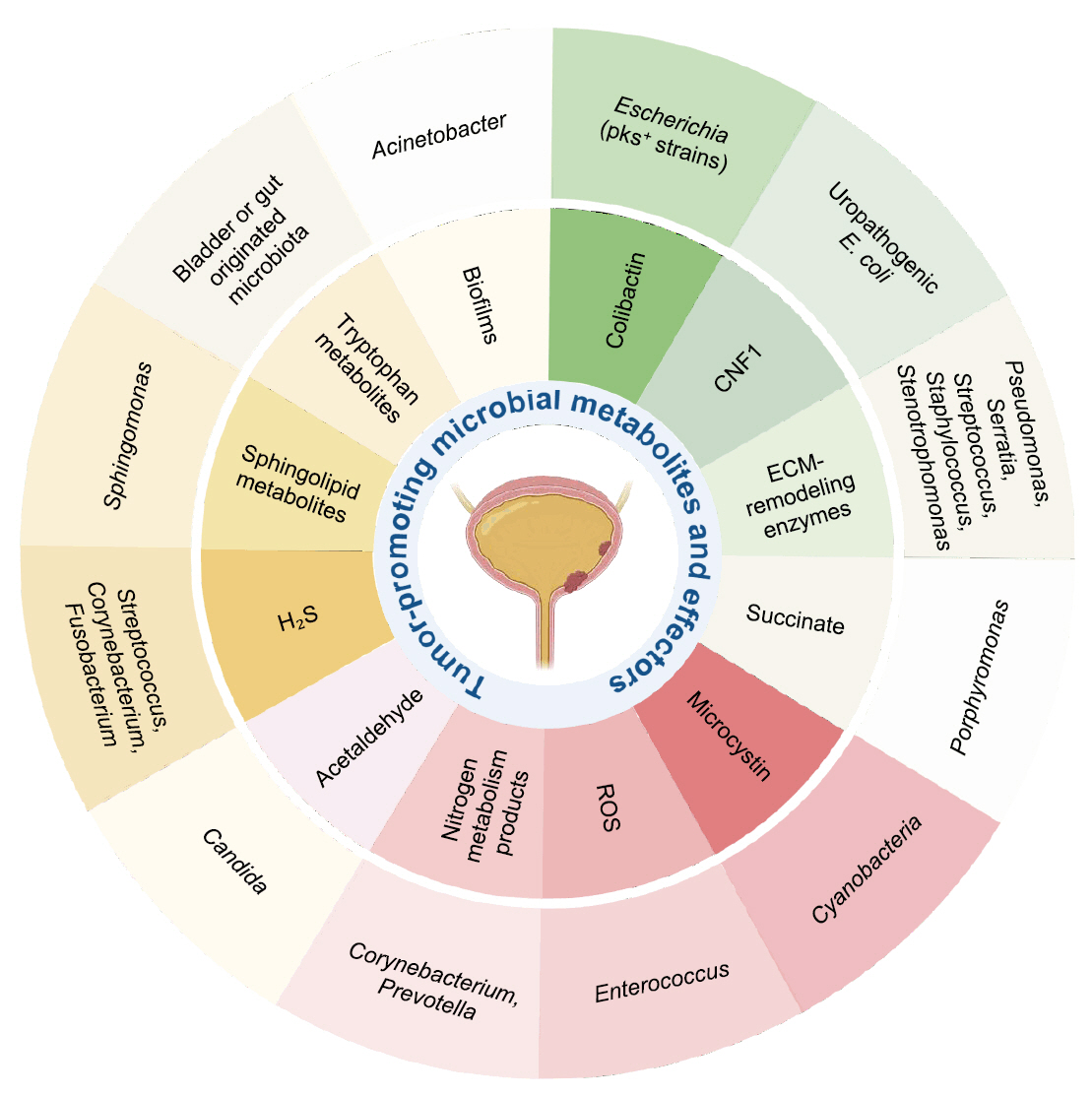
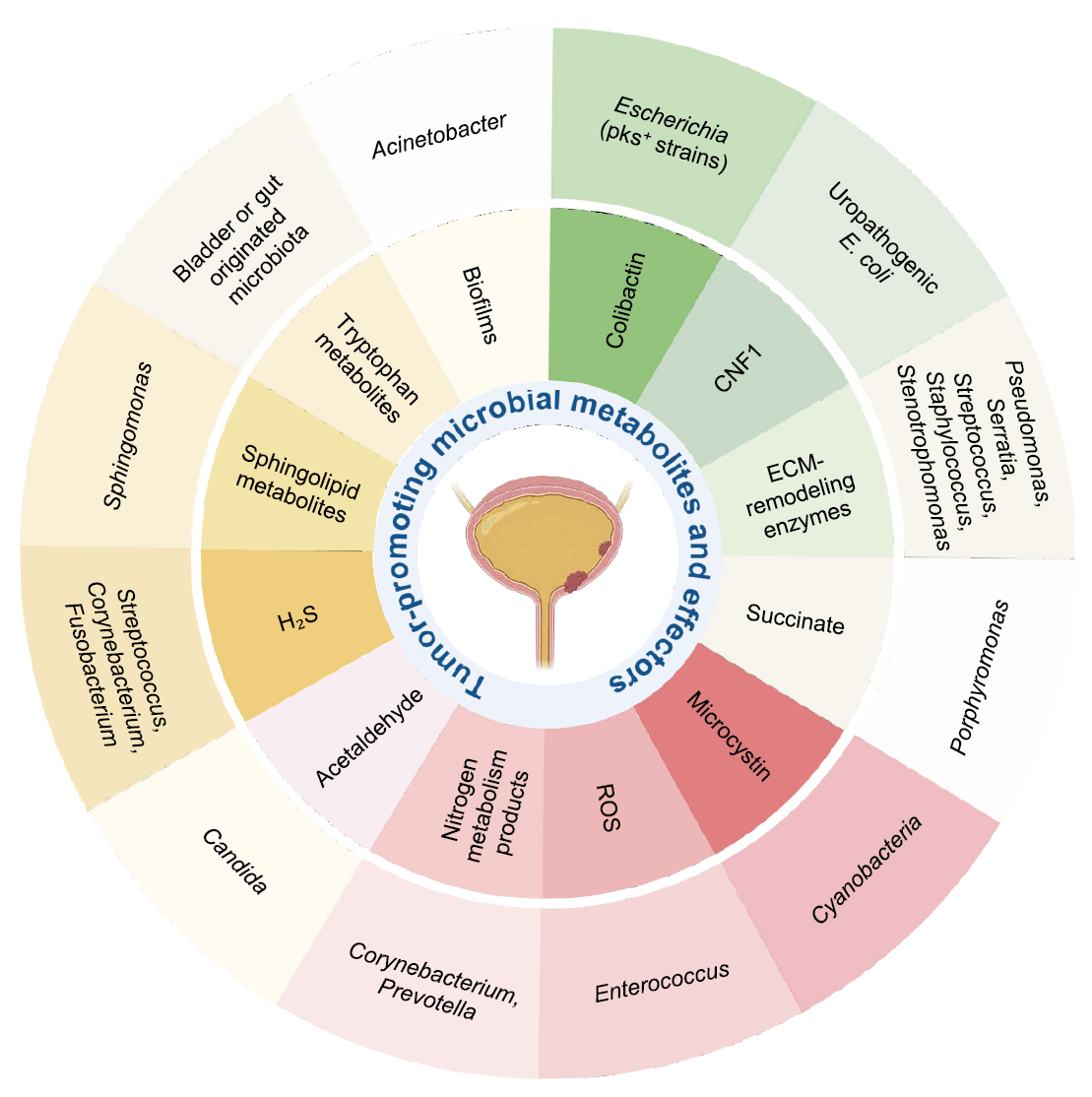
Microbial Metabolites and Bladder Cancer Progression
The Role of the Microbiota–Metabolite–Immunity Axis in Bladder Cancer and Precision Therapeutic Strategies
Conclusion and Future Perspectives
Acknowledgments
We thank our collaborators for reviewing this manuscript. This work was supported by the National Research Foundation of Korea grant (RS-2023-00221182 and RS-2025-00515944) and the internal grant of Korea University.
Author Contributions
Thu Anh Trần: Conceptualization, Writing - Original Draft
Ho Young Lee: Writing - Review & Editing
Hae Woong Choi: Conceptualization, Writing - Original Draft, Funding acquisition.
Conflict of Interest
The authors declare no competing interests.
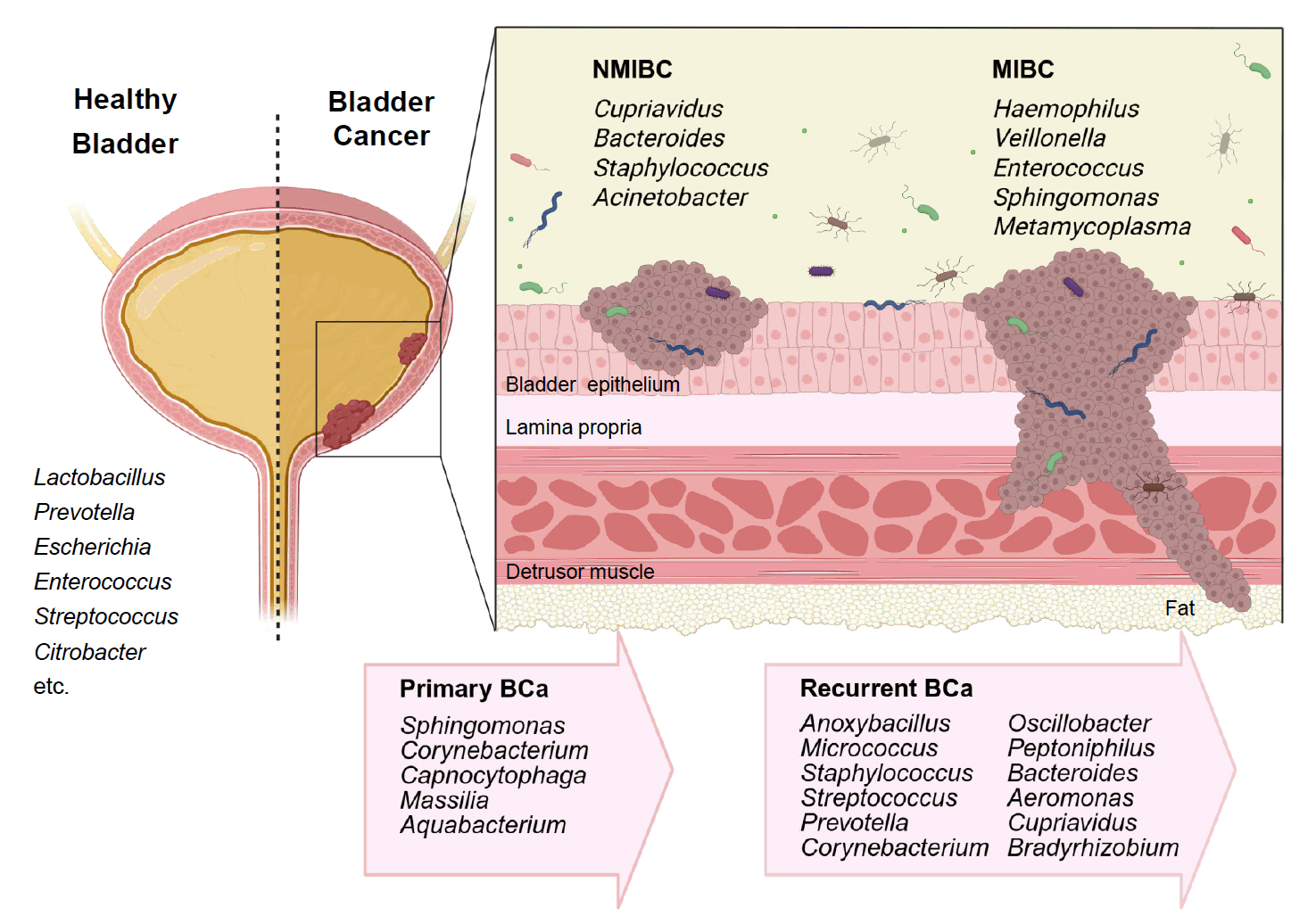
| Specimen (Study/Year) | Case/Control | NMIBC/MIBC | Gender | Enriched genera in BC |
|---|---|---|---|---|
| Midstream urine, Bladder tissue (Bučević Popović et al., 2018) | 12/11 | Primary NMIBC (10)/Recurrent NMIBC (2) | M | Fusobacterium, Actinobaculum, Facklamia, Campylobacter, Subdoligranulum |
| Midstream urine (Wu et al., 2018) | 31/18 | 26/5 | M | Acinetobacter, Anaerococcus, Rubrobacter, Sphingobacterium, Atopostipes, Geobacillus, Herbaspirillum, Porphyrobacter |
| Bacteroides were particularly enriched in high-risk patients | ||||
| Bladder tissue (Liu et al., 2019) | 22/0 | 5/17 | M | Cupriavidus spp., unclassified Brucellaceae, Acinetobacter, Escherichia-Shigella, Sphingomonas, Pelomonas, Ralstonia, Anoxybacillus, Geobacillus |
| Midstream urine (Zeng et al., 2020) | 62/19 | 51/11 | M | Recurrence NMIBC: Anoxybacillus, Micrococcus, Staphylococcus, Streptococcus, Prevotella, Corynebacterium_1, Oscillobacter, Peptoniphilus, Bacteroides |
| Transurethral resectoscopy urine, Bladder tissue (Mansour et al., 2020) | 10/0 | 6/4 | F, M | Urine samples: Lactobacillus, Corynebacterium, Streptococcus, Staphylococcus |
| Tissue samples: Bacteroides, Akkermansia, Klebsiella, Enterobacter, Clostridium sensu stricto. | ||||
| Midstream urine, Tissue (Pederzoli et al., 2020) | 49/59 | Mixed | F, M | Urine samples: Klebsiella (only in female) |
| Tissue samples: Burkholderia (both in male and female) | ||||
| Urine (midstream or catheter) (Hussein et al., 2021) | 43/10 | 29/14 | F, M | Both type: Actinomyces, Achromobacter, Brevibacterium, Brucella |
| NMIBC: Cupriavidus | ||||
| MIBC: Haemophilus, Veillonella | ||||
| BCG responder (NMIBC): Serratia, Brochothrix, Negativicoccus, Escherichia-Shigella, Pseudomonas | ||||
| Urine (midstream or catheter), bladder washout (Oresta et al., 2021) | 51/10 | Mixed | F, M | Midstream urines: Streptococcus, Enterococcus, Corynebacterium, Fusobacterium |
| Bladder washouts: Burkholderiaceae | ||||
| Catheterized urines: Veillonella, Corynebacterium | ||||
| Bladder tissue (Parra-Grande et al., 2022) | 32/0 | Mixed | F, M | Bacteroidetes, Escherichia‑Shigella, Enterococcus, Barnesiella, Parabacteroides, Prevotella, Alistipes, Staphylococcus |
| Midstream urine (Chorbińska et al., 2023) | 18/7 | Mixed | F, M | Howardellagenus, Streptococcus anginosus. |
| *Lactobacillus was more frequent in BCG-treated patients. | ||||
| Bladder tumor tissue (Sun et al., 2023) | 22/0 | 7/15 | F, M | Both type: Ralstonia, Cutibacterium |
| NMIBC: Bacteroides, Staphylococcus, Acinetobacter | ||||
| MBIC: Enterococcus, Sphingomonas, Metamycoplasma | ||||
| Catheterized urine (Heidrich et al., 2024) | 32/41 | 32/0 | M | No significant differences in microbiota composition (NMIBC vs. controls) |
| * Association of Lactobacillus, Streptococcus, Cutibacterium with better BCG response | ||||
| Midstream urine, bladder tissue (Bilski et al., 2024) | 41/0 | 22/19 | F, M | Male: Campylobacter, Sphingobium, Haemophilus, Aeribacillus, Peptococcus, Alcaligenes, Actinomyces, Pseudomonas, Acinetobacter, Proteus |
| Female: Salmonella, Romboutsia, Enterobacter | ||||
| Midstream urine (Sheng et al., 2025b) | 170/0 | Mixed | F, M | Primary BCa: Sphingomonas, Corynebacterium, Capnocytophaga, Massilia, Aquabacterium |
| Primary (39) vs Recurrent (39) | Recurrent BCa: Aeromonas, Cupriavidus, Bradyrhizobium | |||
| Midstream urine (Ginwala et al., 2025) | 55/13 | Mixed | F, M | Enterobacteriales, Flavobacterium, Varicubaculum, Facklamia |
| Chemotherapy non-responders: Granulicatella, Proteus | ||||
| Chemotherapy responders: Enterococcus faecalis |
| Metabolite/Effector | Representative microbial taxa | Mechanistic effect in bladder cancer |
|---|---|---|
| Colibactin (Chagneau et al., 2021; Dziubańska-Kusibab et al., 2020; Nougayrède et al., 2006) | Escherichia coli (pks+ strains) | DNA alkylation, double-strand breaks, mutational signature |
| Cytotoxic Necrotizing Factor 1 (CNF1) (Guo et al., 2020) | Uropathogenic E. coli | RhoC–HIF1α–VEGF pathway activation → angiogenesis |
| ECM-remodeling enzymes (Alfano et al., 2016; DuMont and Cianciotto, 2017; Odunitan et al., 2024) | Pseudomonas, Serratia, Staphylococcus, Stenotrophomonas | ECM degradation, junction disruption, enhanced invasion |
| Succinate (Crooks et al., 2021; Nardelli et al., 2024) | Porphyromonas | HIF-1α stabilization, ECM remodeling, ROS generation |
| Microcystin (Mansour et al., 2020; Svirčev et al., 2010) | Cyanobacteria | DNA damage, enhanced invasiveness, environmental influence |
| Reactive Oxygen Species (ROS) (Huycke et al., 2002; Williamson et al., 2022) | Enterococcus | DNA damage, inflammation, epithelial injury |
| Nitrogen metabolism products (Sheng et al., 2025a; Soriano and Tauch, 2008) | Corynebacterium, Prevotella | pH shift, nitrogen metabolite buildup, chronic inflammation |
| Acetaldehyde (Lao et al., 2021; Nieminen et al., 2009) | Candida | DNA damage, chronic inflammation |
| Hydrogen sulfide (H₂S) (Wang et al., 2023) | Streptococcus,Corynebacterium, Fusobacterium | NF-κB/MAPK activation, immune modulation, autophagy |
| Sphingolipid metabolites (Mlynarczyk et al., 2024; Ponnusamy et al., 2012) | Sphingomonas | Cell proliferation, angiogenesis, immune modulation |
| Tryptophan metabolites (Nizioł et al., 2023; Opitz et al., 2011) | Bladder or gut originated microbiota | AHR activation, immunosuppression |
| Biofilms (Johnson et al., 2015; Nadler et al., 2021) | Acinetobacter | ECM remodeling, immune evasion, chronic inflammation |
- Al Awamlh BAH, Chang SS. 2023. Novel therapies for high-risk non-muscle invasive bladder cancer. Curr Oncol Rep. 25: 83–91. ArticlePubMedPDF
- Alfano M, Canducci F, Nebuloni M, Clementi M, Montorsi F, et al. 2016. The interplay of extracellular matrix and microbiome in urothelial bladder cancer. Nat Rev Urol. 13: 77–90. ArticlePubMedPDF
- Amieva M, Peek RM Jr. 2016. Pathobiology of Helicobacter pylori-induced gastric cancer. Gastroenterology. 150: 64–78. ArticlePubMed
- Bilski KZ, Żeber-Lubecka N, Kulecka M, Dąbrowska M, Bałabas A, et al. 2024. Microbiome sex-related diversity in non-muscle-invasive urothelial bladder cancer. Curr Issues Mol Biol. 46: 3595–3609. ArticlePubMedPMC
- Boban T, Milić Roje B, Knezović D, Jerončić A, Šošić H, et al. 2025. Urinary microbiota changes among NMIBC patients during BCG therapy: comparing BCG responders and non-responders. Front Cell Infect Microbiol. 15: 1479795.ArticlePubMedPMC
- Bray F, Laversanne M, Sung H, Ferlay J, Siegel RL, et al. 2024. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 74: 229–263. ArticlePubMed
- Bučević Popović V, Šitum M, Chow CET, Chan LS, Roje B, et al. 2018. The urinary microbiome associated with bladder cancer. Sci Rep. 8: 12157.ArticlePubMedPMC
- Bukavina L, Isali I, Ginwala R, Sindhani M, Calaway A, et al. 2023. Global meta-analysis of urine microbiome: colonization of polycyclic aromatic hydrocarbon-degrading bacteria among bladder cancer patients. Eur Urol Oncol. 6: 190–203. ArticlePubMed
- Buratti FM, Scardala S, Funari E, Testai E. 2011. Human glutathione transferases catalyzing the conjugation of the hepatoxin microcystin-LR. Chem Res Toxicol. 24: 926–933. ArticlePubMed
- Carlström M, Moretti CH, Weitzberg E, Lundberg JO. 2020. Microbiota, diet and the generation of reactive nitrogen compounds. Free Radic Biol Med. 161: 321–325. ArticlePubMed
- Chagneau CV, Massip C, Bossuet-Greif N, Fremez C, Motta JP, et al. 2021. Uropathogenic E. coli induces DNA damage in the bladder. PLoS Pathog. 17: e1009310. ArticlePubMedPMC
- Chorbińska J, Krajewski W, Nowak Ł, Bardowska K, Żebrowska-Różańska P, et al. 2023. Is the urinary and gut microbiome associated with bladder cancer? Clin Med Insights Oncol. 17: 11795549231206796.ArticlePubMedPMC
- Colella M, Topi S, Palmirotta R, D’Agostino D, Charitos IA, et al. 2023. An overview of the microbiota of the human urinary tract in health and disease: current issues and perspectives. Life. 13: 1486.ArticlePubMedPMC
- Crooks TA, Madison JD, Walsh DM, Herbert WG, Jeraldo PR, et al. 2021. Porphyromonas somerae invasion of endometrial cancer cells. Front Microbiol. 12: 674835.ArticlePubMedPMC
- Cumberbatch MGK, Jubber I, Black PC, Esperto F, Figueroa JD, et al. 2018. Epidemiology of bladder cancer: a systematic review and contemporary update of risk factors in 2018. Eur Urol. 74: 784–795. ArticlePubMed
- de Martel C, Ferlay J, Franceschi S, Vignat J, Bray F, et al. 2012. Global burden of cancers attributable to infections in 2008: a review and synthetic analysis. Lancet Oncol. 13: 607–615. ArticlePubMed
- DuMont AL, Cianciotto NP. 2017. Stenotrophomonas maltophilia serine protease StmPr1 induces matrilysis, anoikis, and protease-activated receptor 2 activation in human lung epithelial cells. Infect Immun. 85: e00544-17.ArticlePubMedPMCLink
- Dziubańska-Kusibab PJ, Berger H, Battistini F, Bouwman BAM, Iftekhar A, et al. 2020. Colibactin DNA-damage signature indicates mutational impact in colorectal cancer. Nat Med. 26: 1063–1069. ArticlePubMedPDF
- Fankhauser CD, Mostafid H. 2018. Genetic polymorphisms may explain association between alcohol consumption and bladder cancer risk in East Asian men. Transl Androl Urol. 7: S252–S254. ArticlePubMedPMC
- Fouts DE, Pieper R, Szpakowski S, Pohl H, Knoblach S, et al. 2012. Integrated next-generation sequencing of 16S rDNA and metaproteomics differentiate the healthy urine microbiome from asymptomatic bacteriuria in neuropathic bladder associated with spinal cord injury. J Transl Med. 10: 174.ArticlePubMedPMCPDF
- Gagnaire A, Nadel B, Raoult D, Neefjes J, Gorvel JP. 2017. Collateral damage: insights into bacterial mechanisms that predispose host cells to cancer. Nat Rev Microbiol. 15: 109–128. ArticlePubMedPDF
- Geller LT, Barzily-Rokni M, Danino T, Jonas OH, Shental N, et al. 2017. Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science. 357: 1156–1160. ArticlePubMedPMC
- Ginwala R, Bukavina L, Sindhani M, Nachman E, Peri S, et al. 2025. Bladder cancer microbiome and its association with chemoresponse. Front Oncol. 15: 1506319.ArticlePubMedPMC
- Guo Y, Wang J, Zhou K, Lv J, Wang L, et al. 2020. Cytotoxic necrotizing factor 1 promotes bladder cancer angiogenesis through activating RhoC. FASEB J. 34: 7927–7940. ArticlePubMedLink
- Heidrich V, Mariotti ACH, Inoue LT, Coser EM, dos Santos EX, et al. 2024. The bladder microbiota is not significantly altered by intravesical BCG therapy. Urol Oncol. 42: 22.Article
- Huang J, Zhang L, Wan D, Zhou L, Zheng S, et al. 2021. Extracellular matrix and its therapeutic potential for cancer treatment. Signal Transduct Target Ther. 6: 153.ArticlePubMedPMCPDF
- Hussein AA, Elsayed AS, Durrani M, Jing Z, Iqbal U, et al. 2021. Investigating the association between the urinary microbiome and bladder cancer: an exploratory study. Urol Oncol. 39: 370.Article
- Huycke MM, Abrams V, Moore DR. 2002. Enterococcus faecalis produces extracellular superoxide and hydrogen peroxide that damages colonic epithelial cell DNA. Carcinogenesis. 23: 529–536. ArticlePubMed
- Johnson CH, Dejea CM, Edler D, Hoang LT, Santidrian AF, et al. 2015. Metabolism links bacterial biofilms and colon carcinogenesis. Cell Metab. 21: 891–897. ArticlePubMedPMC
- King A, Selak MA, Gottlieb E. 2006. Succinate dehydrogenase and fumarate hydratase: linking mitochondrial dysfunction and cancer. Oncogene. 25: 4675–4682. ArticlePubMedPDF
- Kustrimovic N, Bilato G, Mortara L, Baci D. 2024. The urinary microbiome in health and disease: relevance for bladder cancer. Int J Mol Sci. 25: 1732.ArticlePubMedPMC
- Lao Y, Li X, He L, Guan X, Li R, et al. 2021. Association between alcohol consumption and risk of bladder cancer: a dose-response meta-analysis of prospective cohort studies. Front Oncol. 11: 696676.ArticlePubMedPMC
- Lewis DA, Brown R, Williams J, White P, Jacobson SK, et al. 2013. The human urinary microbiome; bacterial DNA in voided urine of asymptomatic adults. Front Cell Infect Microbiol. 3: 41.ArticlePubMedPMC
- Li WT, Iyangar AS, Reddy R, Chakladar J, Bhargava V, et al. 2021. The bladder microbiome is associated with epithelial-mesenchymal transition in muscle invasive urothelial bladder carcinoma. Cancers. 13: 3649.ArticlePubMedPMC
- Li N, Wang L, Yang Q, Li F, Shi Z, et al. 2025. Identification and evaluation of the urinary microbiota associated with bladder cancer. Cancer Innov. 4: e70012. ArticlePubMedPMC
- Lindberg U, Hanson L, Jodal U, Lidin-Janson G, Lincoln K, et al. 1975. Asymptomatic bacteriuria in schoolgirls: II. Differences in Escherichia coli causing asymptomatic and symptomatic bacteriuria. Acta Paediatr Scand. 64: 432–436. ArticlePubMed
- Liu F, Liu A, Lu X, Zhang Z, Xue Y, et al. 2019. Dysbiosis signatures of the microbial profile in tissue from bladder cancer. Cancer Med. 8: 6904–6914. ArticlePubMedPMCLink
- Lundberg JO, Weitzberg E. 2013. Biology of nitrogen oxides in the gastrointestinal tract. Gut. 62: 616–629. ArticlePubMed
- Mansour B, Monyók Á, Makra N, Gajdács M, Vadnay I, et al. 2020. Bladder cancer-related microbiota: examining differences in urine and tissue samples. Sci Rep. 10: 11042.ArticlePubMedPMCPDF
- McConnell MJ, Actis L, Pachón J. 2013. Acinetobacter baumannii: human infections, factors contributing to pathogenesis and animal models. FEMS Microbiol Rev. 37: 130–155. ArticlePubMed
- Mlynarczyk G, Miklosz A, Chabowski A, Baranowski M. 2024. Urothelial urinary bladder cancer is characterized by stage-dependent aberrations in metabolism of bioactive sphingolipids. Int J Mol Sci. 25: 11889.ArticlePubMedPMC
- Modena BD, Milam R, Harrison F, Cheeseman JA, Abecassis MM, et al. 2017. Changes in urinary microbiome populations correlate in kidney transplants with interstitial fibrosis and tubular atrophy documented in early surveillance biopsies. Am J Transplant. 17: 712–723. ArticlePubMedLink
- Multinu F, Chen J, Madison JD, Torres M, Casarin J, et al. 2020. Analysis of DNA methylation in endometrial biopsies to predict risk of endometrial cancer. Gynecol Oncol. 156: 682–688. ArticlePubMedPMC
- Murphy EC, Frick IM. 2013. Gram-positive anaerobic cocci - commensals and opportunistic pathogens. FEMS Microbiol Rev. 37: 520–553. ArticlePubMed
- Nadler N, Kvich L, Bjarnsholt T, Jensen JB, Gögenur I, et al. 2021. The discovery of bacterial biofilm in patients with muscle invasive bladder cancer. APMIS. 129: 265–270. ArticlePubMedLink
- Nardelli C, Aveta A, Pandolfo SD, Tripodi L, Russo F, et al. 2024. Microbiome profiling in bladder cancer patients using the first-morning urine sample. Eur Urol Open Sci. 59: 18–26. ArticlePubMed
- Naskar M, Parekh VP, Abraham MA, Alibasic Z, Kim MJ, et al. 2023. α-Hemolysin promotes uropathogenic E. coli persistence in bladder epithelial cells via abrogating bacteria-harboring lysosome acidification. PLoS Pathog. 19: e1011388. ArticlePubMedPMC
- Nieminen MT, Uittamo J, Salaspuro M, Rautemaa R. 2009. Acetaldehyde production from ethanol and glucose by non-Candida albicans yeasts in vitro. Oral Oncol. 45: e245–e248. ArticlePubMed
- Nizioł J, Ossoliński K, Płaza-Altamer A, Kołodziej A, Ossolińska A, et al. 2023. Untargeted urinary metabolomics for bladder cancer biomarker screening with ultrahigh-resolution mass spectrometry. Sci Rep. 13: 9802.ArticlePubMedPMC
- Nougayrède JP, Homburg S, Taieb F, Boury M, Brzuszkiewicz E, et al. 2006. Escherichia coli induces DNA double-strand breaks in eukaryotic cells. Science. 313: 848–851. ArticlePubMed
- Odunitan TT, Apanisile BT, Akinboade MW, Abdulazeez WO, Oyaronbi AO, et al. 2024. Microbial mysteries: Staphylococcus aureus and the enigma of carcinogenesis. Microb Pathog. 194: 106831.ArticlePubMed
- Opitz CA, Litzenburger UM, Sahm F, Ott M, Tritschler I, et al. 2011. An endogenous tumour-promoting ligand of the human aryl hydrocarbon receptor. Nature. 478: 197–203. ArticlePubMedPDF
- Oresta B, Braga D, Lazzeri M, Frego N, Saita A, et al. 2021. The microbiome of catheter collected urine in males with bladder cancer according to disease stage. J Urol. 205: 86–93. ArticlePubMed
- Parhi L, Alon-Maimon T, Sol A, Nejman D, Shhadeh A, et al. 2020. Breast cancer colonization by Fusobacterium nucleatum accelerates tumor growth and metastatic progression. Nat Commun. 11: 3259.ArticlePubMedPMCPDF
- Parida S, Sharma D. 2019. The power of small changes: comprehensive analyses of microbial dysbiosis in breast cancer. Biochim Biophys Acta Rev Cancer. 1871: 392–405. ArticlePubMedPMC
- Parra-Grande M, Oré-Arce M, Martinez-Priego L, D’Auria G, Rosselló-Mora R, et al. 2022. Profiling the bladder microbiota in patients with bladder cancer. Front Microbiol. 12: 718776.ArticlePubMedPMC
- Pathoor NN, Ganesh PS, Gopal RK. 2024. Microbiome interactions: Acinetobacter baumannii biofilms as a co-factor in oral cancer progression. World J Microbiol Biotechnol. 40: 398.ArticlePDF
- Pederzoli F, Ferrarese R, Amato V, Locatelli I, Alchera E, et al. 2020. Sex-specific alterations in the urinary and tissue microbiome in therapy-naïve urothelial bladder cancer patients. Eur Urol Oncol. 3: 784–788. Article
- Pleguezuelos-Manzano C, Puschhof J, Rosendahl Huber A, van Hoeck A, Wood HM, et al. 2020. Mutational signature in colorectal cancer caused by genotoxic pks+ E. coli. Nature. 580: 269–273. ArticlePubMedPMC
- Ponnusamy S, Selvam SP, Mehrotra S, Kawamori T, Snider AJ, et al. 2012. Communication between host organism and cancer cells is transduced by systemic sphingosine kinase 1/sphingosine 1-phosphate signalling to regulate tumour metastasis. EMBO Mol Med. 4: 761–775. ArticlePubMedPMCLink
- Ripoll J, Ramos M, Montaño J, Pons J, Ameijide A, et al. 2021. Cancer-specific survival by stage of bladder cancer and factors collected by Mallorca Cancer Registry associated to survival. BMC Cancer. 21: 676.ArticlePubMedPMCPDF
- Russo F, Esposito S, Tripodi L, Pandolfo SD, Aveta A, et al. 2024. Insights into Porphyromonas somerae in bladder cancer patients: urinary detection by ddPCR. Microorganisms. 12: 2049.ArticlePubMedPMC
- Saginala K, Barsouk A, Aluru JS, Rawla P, Padala SA, et al. 2020. Epidemiology of bladder cancer. Med Sci. 8: 15.Article
- Sheng Z, Liu J, Wang M, Chen X, Xu J, et al. 2025a. Exploring bladder cancer through urinary microbiota: innovative “urinetypes” classification and establishment of a diagnostic model. J Transl Med. 23: 809.ArticlePDF
- Sheng Z, Xu J, Wang M, Xu X, Zhu J, et al. 2025b. The role of urinary microbiota in primary and recurrent bladder cancer: insights from a propensity score matching study. BMC Cancer. 25: 468.ArticlePDF
- Song CH, Kim YH, Naskar M, Hayes BW, Abraham MA, et al. 2022. Lactobacillus crispatus limits bladder uropathogenic E. coli infection by triggering a host type I interferon response. Proc Natl Acad Sci USA. 119: e2117904119. ArticlePubMedPMC
- Soriano F, Tauch A. 2008. Microbiological and clinical features of Corynebacterium urealyticum: urinary tract stones and genomics as the Rosetta Stone. Clin Microbiol Infect. 14: 632–643. ArticlePubMed
- Su X, Gao Y, Yang R. 2022. Gut microbiota-derived tryptophan metabolites maintain gut and systemic homeostasis. Cells. 11: 2296.ArticlePubMedPMC
- Su W, Shi J, Zhao Y, Yan F, Lei L, et al. 2020. Porphyromonas gingivalis triggers inflammatory responses in periodontal ligament cells by succinate-succinate dehydrogenase-HIF-1α axis. Biochem Biophys Res Commun. 522: 184–190. ArticlePubMed
- Sun JX, Xia QD, Zhong XY, Liu Z, Wang SG. 2023. The bladder microbiome of NMIBC and MIBC patients revealed by 2bRAD-M. Front Cell Infect Microbiol. 13: 1182322.ArticlePubMedPMC
- Svirčev Z, Baltić V, Gantar M, Juković M, Stojanović D, et al. 2010. Molecular aspects of microcystin-induced hepatotoxicity and hepatocarcinogenesis. J Environ Sci Health C Environ Carcinog Ecotoxicol Rev. 28: 39–59. ArticlePubMed
- Tan WS, Rodney S, Lamb B, Feneley M, Kelly J. 2016. Management of non-muscle invasive bladder cancer: a comprehensive analysis of guidelines from the United States, Europe and Asia. Cancer Treat Rev. 47: 22–31. ArticlePubMed
- Vermeulen SH, Hanum N, Grotenhuis AJ, Castaño-Vinyals G, van der Heijden AG, et al. 2015. Recurrent urinary tract infection and risk of bladder cancer in the Nijmegen bladder cancer study. Br J Cancer. 112: 594–600. ArticlePubMedPDF
- Wang Y, Chang Q, Li Y. 2018. Racial differences in urinary bladder cancer in the United States. Sci Rep. 8: 12521.ArticlePubMedPMCPDF
- Wang N, Fang JY. 2023. Fusobacterium nucleatum, a key pathogenic factor and microbial biomarker for colorectal cancer. Trends Microbiol. 31: 159–172. ArticlePubMed
- Wang M, Wang Z, Lessing DJ, Guo M, Chu W. 2023. Fusobacterium nucleatum and its metabolite hydrogen sulfide alter gut microbiota composition and autophagy process and promote colorectal cancer progression. Microbiol Spectr. 11: e02292-23.ArticlePubMedPMCLink
- Wang F, Wu H, Fan M, Yu R, Zhang Y, et al. 2020. Sodium butyrate inhibits migration and induces AMPK-mTOR pathway-dependent autophagy and ROS-mediated apoptosis via the miR-139-5p/Bmi-1 axis in human bladder cancer cells. FASEB J. 34: 4266–4282. ArticlePubMedLink
- Williamson AJ, Jacobson R, van Praagh J, Gaines S, Koo HY, et al. 2022. Enterococcus faecalis promotes a migratory and invasive phenotype in colon cancer cells. Neoplasia. 27: 100787.ArticlePubMedPMC
- Wittmann BM, Stirdivant SM, Mitchell MW, Wulff JE, McDunn JE, et al. 2014. Bladder cancer biomarker discovery using global metabolomic profiling of urine. PLoS One. 9: e115870. ArticlePubMedPMC
- Wu C, Wei X, Huang Z, Zheng Z, Zhang W, et al. 2024. Urinary microbiome dysbiosis is associated with an inflammatory environment and perturbed fatty acids metabolism in the pathogenesis of bladder cancer. J Transl Med. 22: 628.ArticlePubMedPMCPDF
- Wu P, Zhang G, Zhao J, Chen J, Chen Y, et al. 2018. Profiling the urinary microbiota in male patients with bladder cancer in China. Front Cell Infect Microbiol. 8: 167.ArticlePubMedPMC
- Zeng J, Zhang G, Chen C, Li K, Wen Y, et al. 2020. Alterations in urobiome in patients with bladder cancer and implications for clinical outcome: a single-institution study. Front Cell Infect Microbiol. 10: 555508.ArticlePubMedPMC
- Zhang W, Yang F, Mao S, Wang R, Chen H, et al. 2023. Bladder cancer-associated microbiota: recent advances and future perspectives. Heliyon. 9: e13012. ArticlePubMedPMC
- Zhou L, Yu H, Chen K. 2002. Relationship between microcystin in drinking water and colorectal cancer. Biomed Environ Sci. 15: 166–171. PubMed
References
Figure & Data
References
Citations

- The infection–microbiome–immunity axis in bladder cancer: mechanistic insights and therapeutic perspectives
Shen Pan, Wanlin Cui, Jiaman Lin, Zhujun Wang, Zhenhua Li, Bitian Liu
Frontiers in Immunology.2026;[Epub] CrossRef
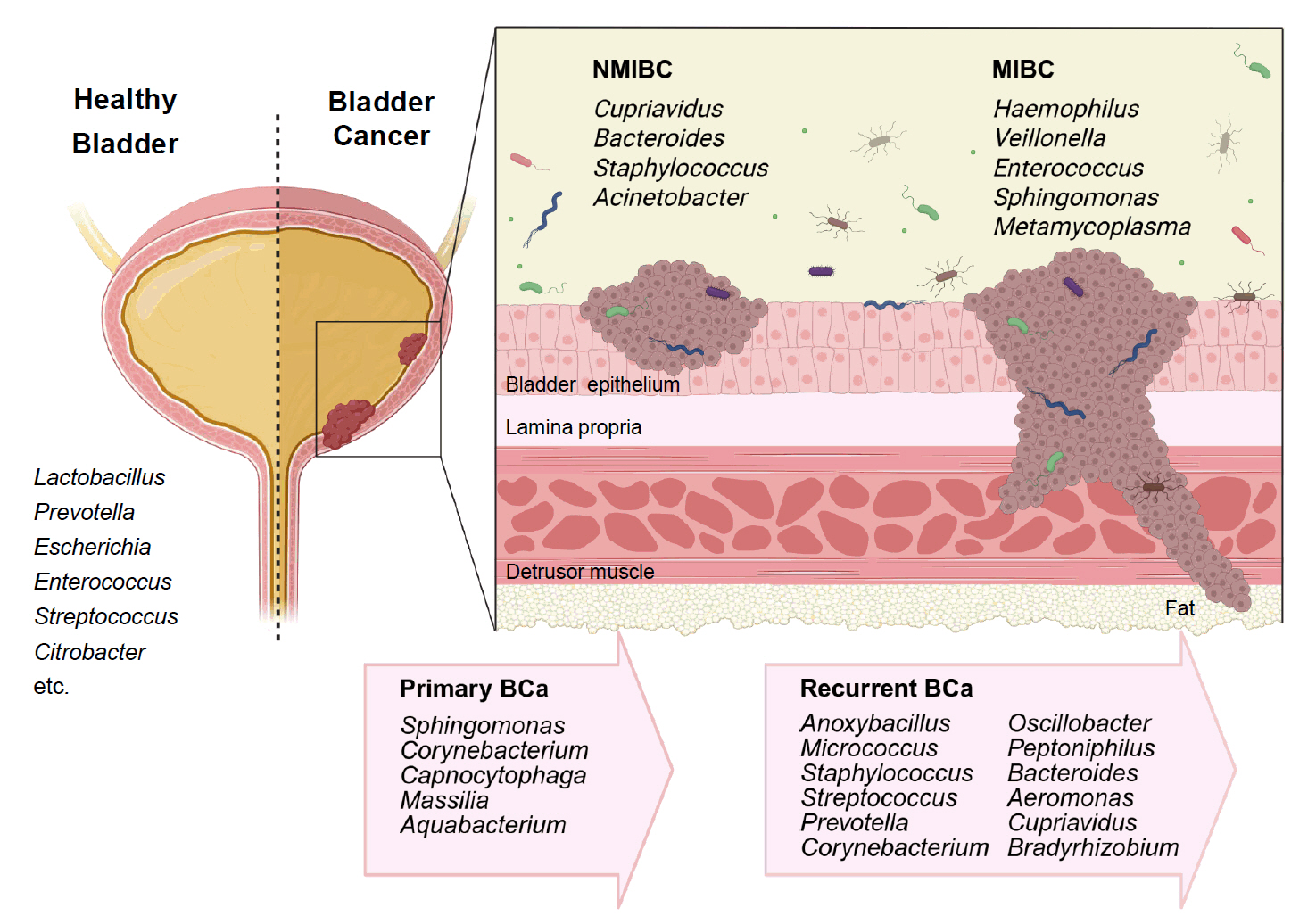
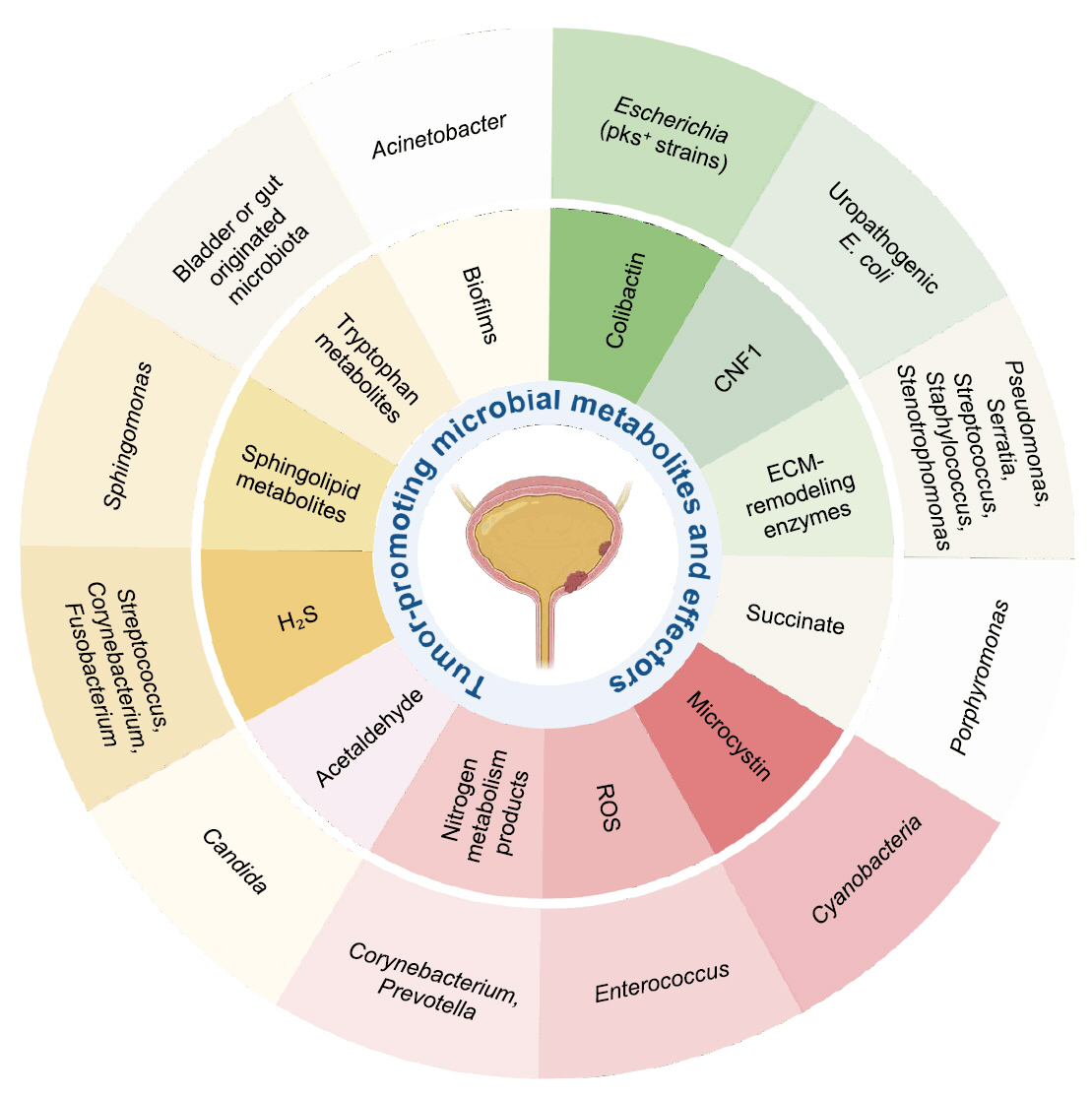
Fig. 1.
Fig. 2.
| Specimen (Study/Year) | Case/Control | NMIBC/MIBC | Gender | Enriched genera in BC |
|---|---|---|---|---|
| Midstream urine, Bladder tissue ( |
12/11 | Primary NMIBC (10)/Recurrent NMIBC (2) | M | Fusobacterium, Actinobaculum, Facklamia, Campylobacter, Subdoligranulum |
| Midstream urine ( |
31/18 | 26/5 | M | Acinetobacter, Anaerococcus, Rubrobacter, Sphingobacterium, Atopostipes, Geobacillus, Herbaspirillum, Porphyrobacter |
| Bacteroides were particularly enriched in high-risk patients | ||||
| Bladder tissue ( |
22/0 | 5/17 | M | Cupriavidus spp., unclassified Brucellaceae, Acinetobacter, Escherichia-Shigella, Sphingomonas, Pelomonas, Ralstonia, Anoxybacillus, Geobacillus |
| Midstream urine ( |
62/19 | 51/11 | M | Recurrence NMIBC: Anoxybacillus, Micrococcus, Staphylococcus, Streptococcus, Prevotella, Corynebacterium_1, Oscillobacter, Peptoniphilus, Bacteroides |
| Transurethral resectoscopy urine, Bladder tissue ( |
10/0 | 6/4 | F, M | Urine samples: Lactobacillus, Corynebacterium, Streptococcus, Staphylococcus |
| Tissue samples: Bacteroides, Akkermansia, Klebsiella, Enterobacter, Clostridium sensu stricto. | ||||
| Midstream urine, Tissue ( |
49/59 | Mixed | F, M | Urine samples: Klebsiella (only in female) |
| Tissue samples: Burkholderia (both in male and female) | ||||
| Urine (midstream or catheter) ( |
43/10 | 29/14 | F, M | Both type: Actinomyces, Achromobacter, Brevibacterium, Brucella |
| NMIBC: Cupriavidus | ||||
| MIBC: Haemophilus, Veillonella | ||||
| BCG responder (NMIBC): Serratia, Brochothrix, Negativicoccus, Escherichia-Shigella, Pseudomonas | ||||
| Urine (midstream or catheter), bladder washout ( |
51/10 | Mixed | F, M | Midstream urines: Streptococcus, Enterococcus, Corynebacterium, Fusobacterium |
| Bladder washouts: Burkholderiaceae | ||||
| Catheterized urines: Veillonella, Corynebacterium | ||||
| Bladder tissue ( |
32/0 | Mixed | F, M | Bacteroidetes, Escherichia‑Shigella, Enterococcus, Barnesiella, Parabacteroides, Prevotella, Alistipes, Staphylococcus |
| Midstream urine ( |
18/7 | Mixed | F, M | Howardellagenus, Streptococcus anginosus. |
| *Lactobacillus was more frequent in BCG-treated patients. | ||||
| Bladder tumor tissue ( |
22/0 | 7/15 | F, M | Both type: Ralstonia, Cutibacterium |
| NMIBC: Bacteroides, Staphylococcus, Acinetobacter | ||||
| MBIC: Enterococcus, Sphingomonas, Metamycoplasma | ||||
| Catheterized urine ( |
32/41 | 32/0 | M | No significant differences in microbiota composition (NMIBC vs. controls) |
| * Association of Lactobacillus, Streptococcus, Cutibacterium with better BCG response | ||||
| Midstream urine, bladder tissue ( |
41/0 | 22/19 | F, M | Male: Campylobacter, Sphingobium, Haemophilus, Aeribacillus, Peptococcus, Alcaligenes, Actinomyces, Pseudomonas, Acinetobacter, Proteus |
| Female: Salmonella, Romboutsia, Enterobacter | ||||
| Midstream urine ( |
170/0 | Mixed | F, M | Primary BCa: Sphingomonas, Corynebacterium, Capnocytophaga, Massilia, Aquabacterium |
| Primary (39) vs Recurrent (39) | Recurrent BCa: Aeromonas, Cupriavidus, Bradyrhizobium | |||
| Midstream urine ( |
55/13 | Mixed | F, M | Enterobacteriales, Flavobacterium, Varicubaculum, Facklamia |
| Chemotherapy non-responders: Granulicatella, Proteus | ||||
| Chemotherapy responders: Enterococcus faecalis |
| Metabolite/Effector | Representative microbial taxa | Mechanistic effect in bladder cancer |
|---|---|---|
| Colibactin ( |
Escherichia coli (pks+ strains) | DNA alkylation, double-strand breaks, mutational signature |
| Cytotoxic Necrotizing Factor 1 (CNF1) ( |
Uropathogenic E. coli | RhoC–HIF1α–VEGF pathway activation → angiogenesis |
| ECM-remodeling enzymes ( |
Pseudomonas, Serratia, Staphylococcus, Stenotrophomonas | ECM degradation, junction disruption, enhanced invasion |
| Succinate ( |
Porphyromonas | HIF-1α stabilization, ECM remodeling, ROS generation |
| Microcystin ( |
Cyanobacteria | DNA damage, enhanced invasiveness, environmental influence |
| Reactive Oxygen Species (ROS) ( |
Enterococcus | DNA damage, inflammation, epithelial injury |
| Nitrogen metabolism products ( |
Corynebacterium, Prevotella | pH shift, nitrogen metabolite buildup, chronic inflammation |
| Acetaldehyde ( |
Candida | DNA damage, chronic inflammation |
| Hydrogen sulfide (H₂S) ( |
Streptococcus,Corynebacterium, Fusobacterium | NF-κB/MAPK activation, immune modulation, autophagy |
| Sphingolipid metabolites ( |
Sphingomonas | Cell proliferation, angiogenesis, immune modulation |
| Tryptophan metabolites ( |
Bladder or gut originated microbiota | AHR activation, immunosuppression |
| Biofilms ( |
Acinetobacter | ECM remodeling, immune evasion, chronic inflammation |
Table 1.
Table 2.
TOP
 MSK
MSK

 ePub Link
ePub Link Cite this Article
Cite this Article