Articles
- Page Path
- HOME > J. Microbiol > Volume 63(11); 2025 > Article
-
Full article
Development of an RT-LAMP−CRISPR/Cas12a assay for rapid and specific detection of Bandavirus dabieense - Bo Seung Song1,†, Yun Hee Baek2,†, Eun-Ha Kim3, Hyeok-Il Kwon4, Ah-Hyeon Kim1, Si-Hyun Lee1, Yu-Bin Son1, Soo-Hyeon Kim1, Min-Suk Song2,*, Young Ki Choi3,*, Su-Jin Park1,5,*
-
Journal of Microbiology 2025;63(11):e2506013.
DOI: https://doi.org/10.71150/jm.2506013
Published online: November 30, 2025
1Division of Applied Life Science, Gyeongsang National University, Jinju 52828, Republic of Korea
2Department of Microbiology, College of Medicine and Medical Research Institute, Chungbuk National University, Cheongju 28644, Republic of Korea
3Center for Study of Emerging and Re-emerging Viruses, Korea Virus Research Institute, Institute for Basic Science (IBS), Daejeon 34126, Republic of Korea
4ChoongAng Vaccine Laboratories, Daejeon 34055, Republic of Korea
5Division of Life Science, College of Natural Sciences, Research Institute of Molecular Alchemy (RIMA), Gyeongsang National University, Jinju 52828, Republic of Korea
-
*Correspondence Min-Suk Song songminsuk@chungbuk.ac.kr
Young Ki Choi choiki55@ibs.re.kr
Su-Jin Park parksujin@gnu.ac.kr - †These authors contributed equally to this work.
© The Microbiological Society of Korea
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/4.0) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
- 1,640 Views
- 107 Download
ABSTRACT
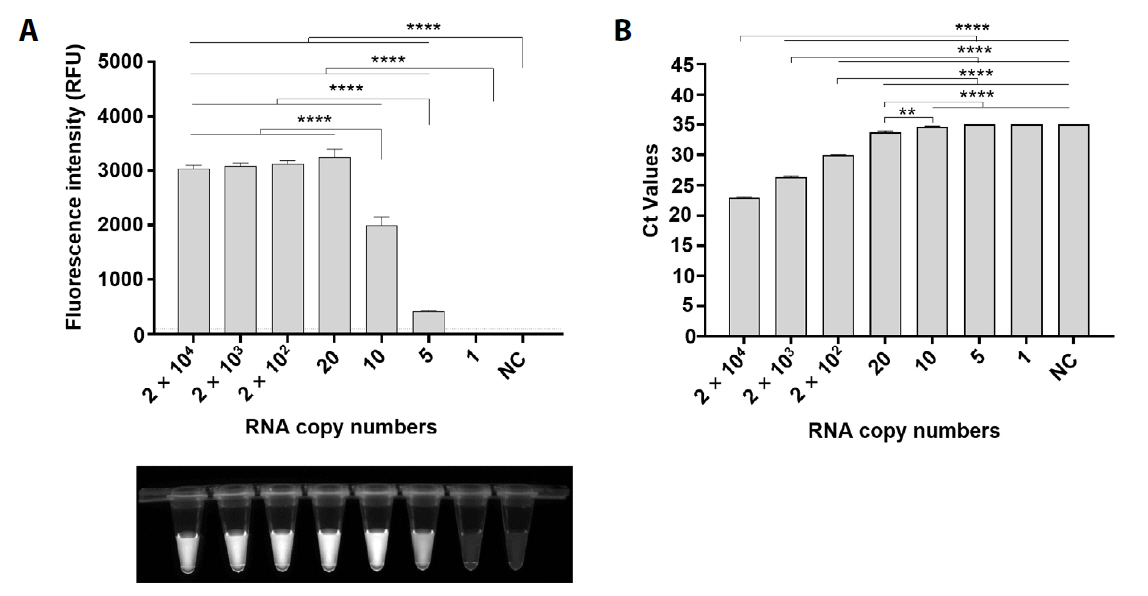
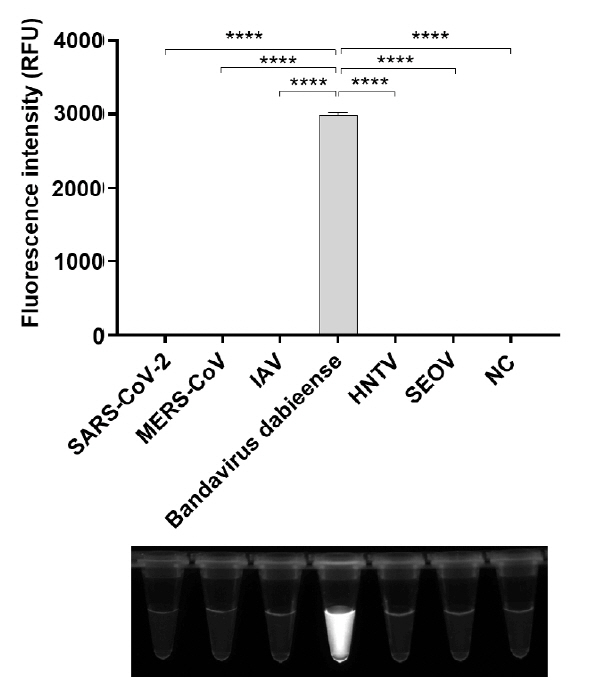
- Bandavirus dabieense, a single-stranded RNA virus, is the causative agent of severe fever with thrombocytopenia syndrome (SFTS), a disease associated with high fatality rates. Early and accurate diagnosis is essential for improving clinical outcomes, particularly given the limited therapeutic options and high mortality rates associated with SFTS. However, while highly sensitive, conventional diagnostic methods such as PCR and qRT-PCR require specialized laboratory facilities and trained personnel, making them impractical for rapid detection in resource-limited settings. To address these challenges, we developed a rapid and highly sensitive assay for Bandavirus dabieense detection by integrating reverse transcription loop-mediated isothermal amplification (RT-LAMP) with CRISPR/Cas12a technology. LAMP primers and guide RNA sequences were designed to target the L gene, ensuring broad detection across viral genotypes. The optimized assay demonstrated a detection limit of 5 RNA copies per reaction, showing more sensitivity than qRT-PCR, and exhibited 100% concordance with qRT-PCR results in clinical samples. Given its speed, accuracy, and field applicability, this LAMP-CRISPR/Cas12a-based assay represents a promising diagnostic tool for early SFTSV detection, particularly in resource-constrained environments where conventional molecular diagnostics are not readily available.
Introduction
Materials and Methods
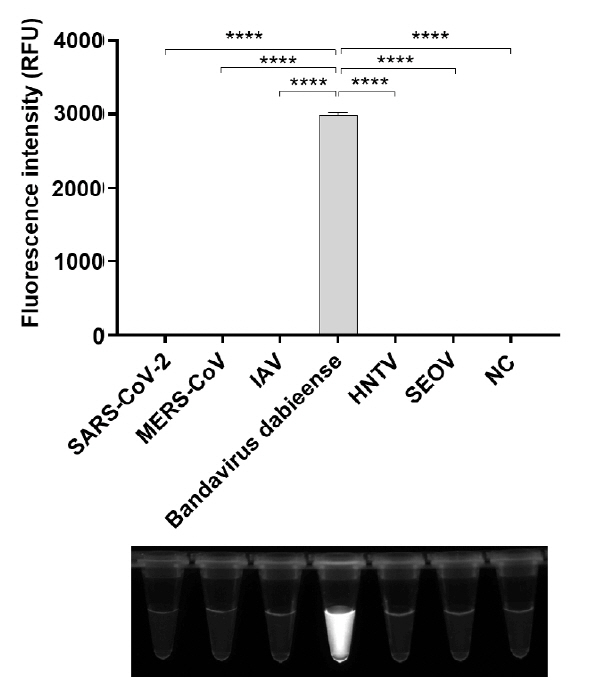
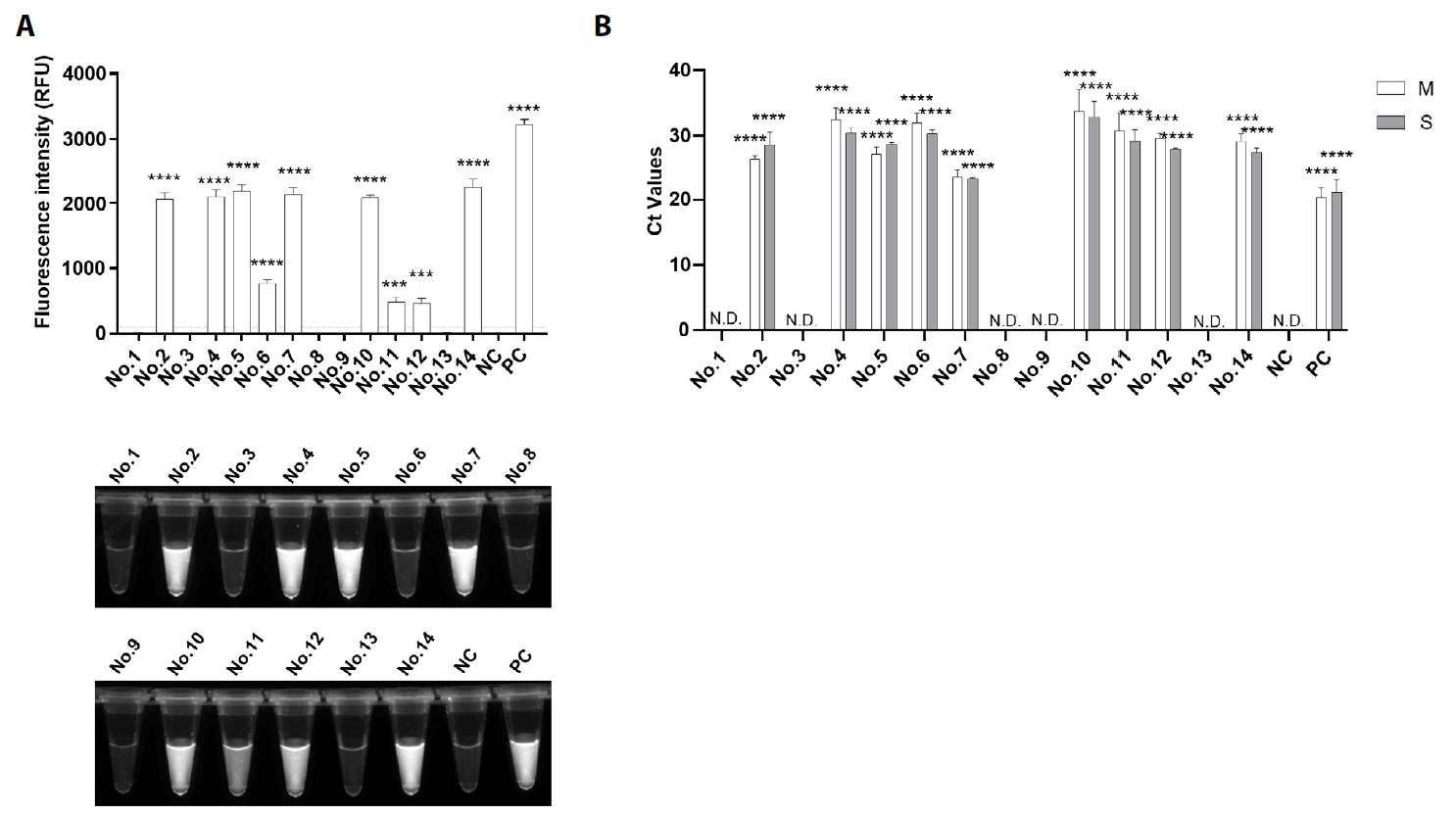
Results
Discussion
Acknowledgments
This research was supported by the National Research Foundation of Korea (NRF) through the MSIT Grant (grant #: NRF-2022R1C1C1004704) and the Learning & Academic Research Institution for Master’s, PhD Students, and Postdocs (LAMP) Program of (grant #: RS-2023-00301974) of the Ministry of Education, as well as by the Korea Institute of Planning and Evaluation for Technology in Food, Agriculture, and Forestry (IPET) through Ministry of Agriculture, Food and Rural Affairs (grant #: RS-2022-IP322088). The Biospecimens and data used in this study were provided by the Biobank of Gyeongsang National University Hospital, a member of Korea Biobank Network.
Conflict of Interest
The authors do not have any competing interests to declare.
Ethical Statements
All human samples used in this study were provided by the Biobank of Gyeongsang National University Hospital, a member of Korea Biobank Network, and were approved by the Chungbuk National University Hospital, in accordance with the approved procedures by the Institutional Review Board of Chungbuk National University Hospital (IRB #. CBNU-202211-BR-0240, CBNU-202402-BR-0289).
| F3 (µM) | B3 (µM) | FIP (µM) | BIP (µM) | LF (µM) | LB (µM) | |
|---|---|---|---|---|---|---|
| A | 0.08 | 0.1 | 0.86 | 0.86 | 0.14 | 0.17 |
| B | 0.18 | 0.2 | 0.86 | 0.86 | 0.14 | 0.17 |
| C | 0.18 | 0.2 | 0.74 | 0.74 | 0.14 | 0.17 |
| D | 0.18 | 0.2 | 0.86 | 0.86 | 0.04 | 0.07 |
- Akagi K, Miyazaki T, Oshima K, Umemura A, Shimada S, et al. 2020. Detection of viral RNA in diverse body fluids in an SFTS patient with encephalopathy, gastrointestinal bleeding and pneumonia: a case report and literature review. BMC Infect Dis. 20: 281.ArticlePubMedPMCPDF
- Aquino-Jarquin G. 2019. CRISPR-Cas14 is now part of the artillery for gene editing and molecular diagnostic. Nanomedicine. 18: 428–431. ArticlePubMed
- Baylis SA, Wallace P, McCulloch E, Niesters HGM, Nübling CM. 2019. Standardization of nucleic acid tests: The approach of the World Health Organization. J Clin Microbiol. 57: e01056-18.ArticlePubMedPMCLink
- Craw P, Balachandran W. 2012. Isothermal nucleic acid amplification technologies for point-of-care diagnostics: a critical review. Lab Chip. 12: 2469–2486. ArticlePubMed
- Deng H, Gao Z. 2015. Bioanalytical applications of isothermal nucleic acid amplification techniques. Anal Chim Acta. 853: 30–45. ArticlePubMed
- Dronina J, Samukaite-Bubniene U, Ramanavicius A. 2022. Towards application of CRISPR-Cas12a in the design of modern viral DNA detection tools (Review). J Nanobiotechnol. 20: 41.ArticlePDF
- Fu Y, Li S, Zhang Z, Man S, Li X, et al. 2016. Phylogeographic analysis of severe fever with thrombocytopenia syndrome virus from Zhoushan islands, China: Implication for transmission across the ocean. Sci Rep. 6: 19563.ArticlePubMedPMCPDF
- Gao X, Sun B, Guan Y. 2019. Pullulan reduces the non-specific amplification of loop-mediated isothermal amplification (LAMP). Anal Bioanal Chem. 411: 1211–1218. ArticlePubMedPDF
- Glökler J, Lim TS, Ida J, Frohme M. 2021. Isothermal amplifications - a comprehensive review on current methods. Crit Rev Biochem Mol Biol. 56: 543–586. ArticlePubMed
- Hu L, Li J, Zhang H, Bian T, Pan J, et al. 2022. Predisposing factors for person-to-person transmission of severe fever with thrombocytopenia syndrome bunyavirus. J Hosp Infect. 123: 174–178. ArticlePubMed
- Huang J, Cook DE. 2022. CRISPR-Cas12a ribonucleoprotein-mediated gene editing in the plant pathogenic fungus Magnaporthe oryzae. STAR Protoc. 3: 101072.ArticlePubMed
- Huang M, Liu S, Xu Y, Li A, Wu W, et al. 2022. CRISPR/Cas12a technology combined with RPA for rapid and portable SFTSV detection. Front Microbiol. 13: 754995.ArticlePubMedPMC
- Jang H, Casel MAB, Jang S, Choi JH, Gil J, et al. 2024. Seasonal dynamics of Haemaphysalis tick species as SFTSV vectors in South Korea. Microbiol Spectr. 12: e00489-24.ArticlePubMedPMCLink
- Jung SI, Kim YE, Yun NR, Kim CM, Kim DM, et al. 2021. Effects of steroid therapy in patients with severe fever with thrombocytopenia syndrome: a multicenter clinical cohort study. PLoS Negl Trop Dis. 15: e0009128. ArticlePubMedPMC
- Kaminski MM, Abudayyeh OO, Gootenberg JS, Zhang F, Collins JJ. 2021. CRISPR-based diagnostics. Nat Biomed Eng. 5: 643–656. ArticlePubMedPDF
- Kim WY, Choi WY, Park SW, Wang EB, Lee WJ, et al. 2015. Nosocomial transmission of severe fever with thrombocytopenia syndrome in Korea. Clin Infect Dis. 60: 1681–1683. ArticlePubMed
- Kim S, Ji S, Koh HR. 2021. CRISPR as a diagnostic tool. Biomolecules. 11: 1162.ArticlePubMedPMC
- Kundu S, Varshney R, Sulabh S. 2024. Exploration of isothermal nucleic acid amplification techniques in the biomedical field. Gene Genome Ed. 7: 100032.Article
- Kwon JS, Kim JY, Jang CY, Son JY, Kim W, et al. 2024. Effect of severe fever with thrombocytopenia syndrome virus genotype on disease severity, viral load, and cytokines in South Korea. Open Forum Infect Dis. 11: ofae508.ArticlePubMedPMCPDF
- Lee K, Seok JH, Kim H, Park S, Lee S, et al. 2023. Genome-informed investigation of the molecular evolution and genetic reassortment of severe fever with thrombocytopenia syndrome virus. PLoS Negl Trop Dis. 17: e0011630. ArticlePubMedPMC
- Li H, Jiang XM, Cui N, Yuan C, Zhang SF, et al. 2021. Clinical effect and antiviral mechanism of T-705 in treating severe fever with thrombocytopenia syndrome. Signal Transduct Target Ther. 6: 145.ArticlePubMedPMCPDF
- Li H, Lu QB, Xing B, Zhang SF, Liu K, et al. 2018. Epidemiological and clinical features of laboratory-diagnosed severe fever with thrombocytopenia syndrome in China, 2011-17: A prospective observational study. Lancet Infect Dis. 18: 1127–1137. ArticlePubMed
- Luo LM, Zhao L, Wen HL, Zhang ZT, Liu JW, et al. 2015. Haemaphysalis longicornis ticks as reservoir and vector of severe fever with thrombocytopenia syndrome virus in China. Emerg Infect Dis. 21: 1770–1776. ArticlePubMedPMC
- Miao D, Dai K, Zhao GP, Li XL, Shi WQ, et al. 2020. Mapping the global potential transmission hotspots for severe fever with thrombocytopenia syndrome by machine learning methods. Emerg Microbes Infect. 9: 817–826. ArticlePubMedPMC
- Murphy J, Bustin SA. 2009. Reliability of real-time reverse-transcription PCR in clinical diagnostics: gold standard or substandard? Expert Rev Mol Diagn. 9: 187–197. ArticlePubMed
- Niemz A, Ferguson TM, Boyle DS. 2011. Point-of-care nucleic acid testing for infectious diseases. Trends Biotechnol. 29: 240–250. ArticlePubMedPMC
- Park D, Kim KW, Kim YI, Casel MAB, Jang H, et al. 2024. Deciphering the evolutionary landscape of severe fever with thrombocytopenia syndrome virus across East Asia. Virus Evol. 10: veae054.ArticlePubMedPMCPDF
- Park BJ, Yoo JR, Heo ST, Kim M, Lee KH, et al. 2022. A CRISPR-Cas12a-based diagnostic method for multiple genotypes of severe fever with thrombocytopenia syndrome virus. PLoS Negl Trop Dis. 16: e0010666. ArticlePubMedPMC
- Reed LJ, Muench H. 1938. A simple method of estimating fifty per cent endpoints. Am J Epidemiol. 27: 493–497. Article
- Senarath KD, Usgodaarachchi RB, Navaratne V, Nagahawatte A, Wijayarathna CD, et al. 2014. Non specific amplification with the LAMP technique in the diagnosis of tuberculosis in Sri Lankan settings. J Tuberc Res. 2: 168–172. ArticleLink
- Shi J, Hu S, Liu X, Yang J, Liu D, et al. 2017. Migration, recombination, and reassortment are involved in the evolution of severe fever with thrombocytopenia syndrome bunyavirus. Infect Genet Evol. 47: 109–117. ArticlePubMed
- Shu J, Tan Q, Huang Z, Zhang T, Ye L, et al. 2025. One-pot one-step detection platform for severe fever with thrombocytopenia syndrome virus via the CRISPR/Cas12a detection system. Virol J. 22: 203.ArticlePubMedPMCPDF
- Takahashi T, Maeda K, Suzuki T, Ishido A, Shigeoka T, et al. 2014. The first identification and retrospective study of severe fever with thrombocytopenia syndrome in Japan. J Infect Dis. 209: 816–827. ArticlePubMed
- Van Der Hofstadt M, l’Helgoualch N, Houot-Cernettig J, Galindo T, Manso T, et al. 2025. Advancing in ovo egg sexing through molecular sex identification of chick embryos using LAMP and RPA assays. Sci Rep. 15: 27722.ArticlePubMedPMC
- Wang Y, Pang B, Wang Z, Tian X, Xu X, et al. 2023. Genomic diversity and evolution analysis of severe fever with thrombocytopenia syndrome in East Asia from 2010 to 2022. Front Microbiol. 14: 1233693.ArticlePubMedPMC
- Wen Y, Ni Z, Hu Y, Wu J, Fang Y, et al. 2024. Multiple genotypes and reassortants of severe fever with thrombocytopenia syndrome virus co-circulating in Hangzhou in southeastern China, 2013-2023. J Med Virol. 96: e70029. ArticlePubMed
- Wu Y, Zhan J, Shan Z, Li Y, Liu Y, et al. 2023. CRISPR-Cas13a-based detection method for avian influenza virus. Front Microbiol. 14: 1288951.ArticlePubMedPMC
- Xu H, Lin G, Chen R, Cai Z, Sun Y, et al. 2024. CRISPR/Cas12b assisted loop-mediated isothermal amplification for easy, rapid and sensitive quantification of chronic HBV DNA in one-pot. Anal Chim Acta. 1310: 342702.ArticlePubMed
- Yan S, Li C, Lan H, Pan D, Wu Y. 2024. Comparison of four isothermal amplification techniques: LAMP, SEA, CPA, and RPA for the identification of chicken adulteration. Food Control. 159: 110302.Article
- Yan L, Zhou J, Zheng Y, Gamson AS, Roembke BT, et al. 2014. Isothermal amplified detection of DNA and RNA. Mol Biosyst. 10: 970–1003. ArticlePubMed
- Yu XJ, Liang MF, Zhang SY, Liu Y, Li JD, et al. 2011. Fever with thrombocytopenia associated with a novel bunyavirus in China. N Engl J Med. 364: 1523–1532. ArticlePubMedPMC
- Yun SM, Park SJ, Kim YI, Park SW, Yu MA, et al. 2020. Genetic and pathogenic diversity of severe fever with thrombocytopenia syndrome virus (SFTSV) in South Korea. JCI Insight. 5: e129531. ArticlePubMedPMC
- Yun SM, Park SJ, Park SW, Choi WY, Jeong HW, et al. 2017. Molecular genomic characterization of tick- and human-derived severe fever with thrombocytopenia syndrome virus isolates from South Korea. PLoS Negl Trop Dis. 11: e0005893. ArticlePubMedPMC
References
Figure & Data
References
Citations

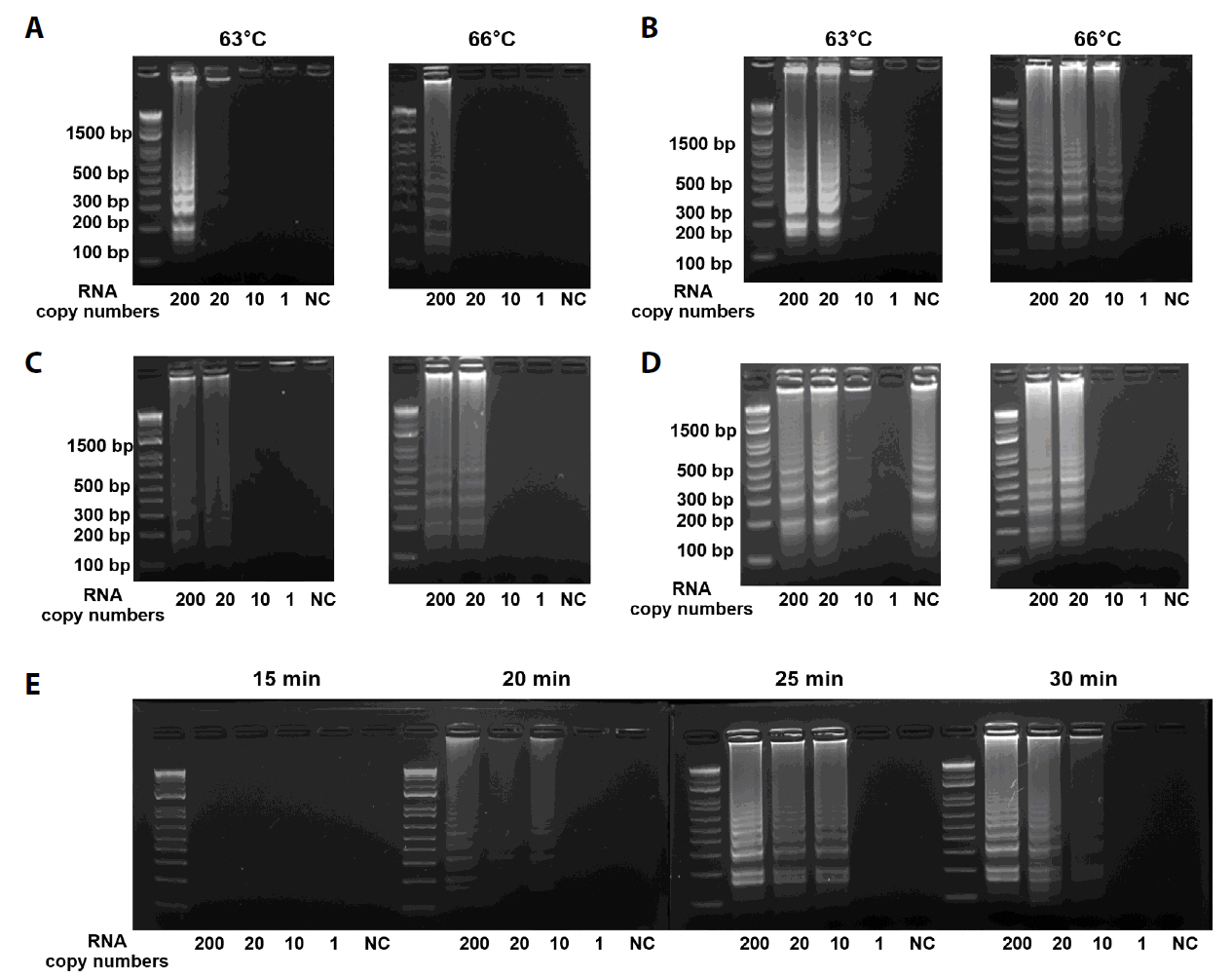
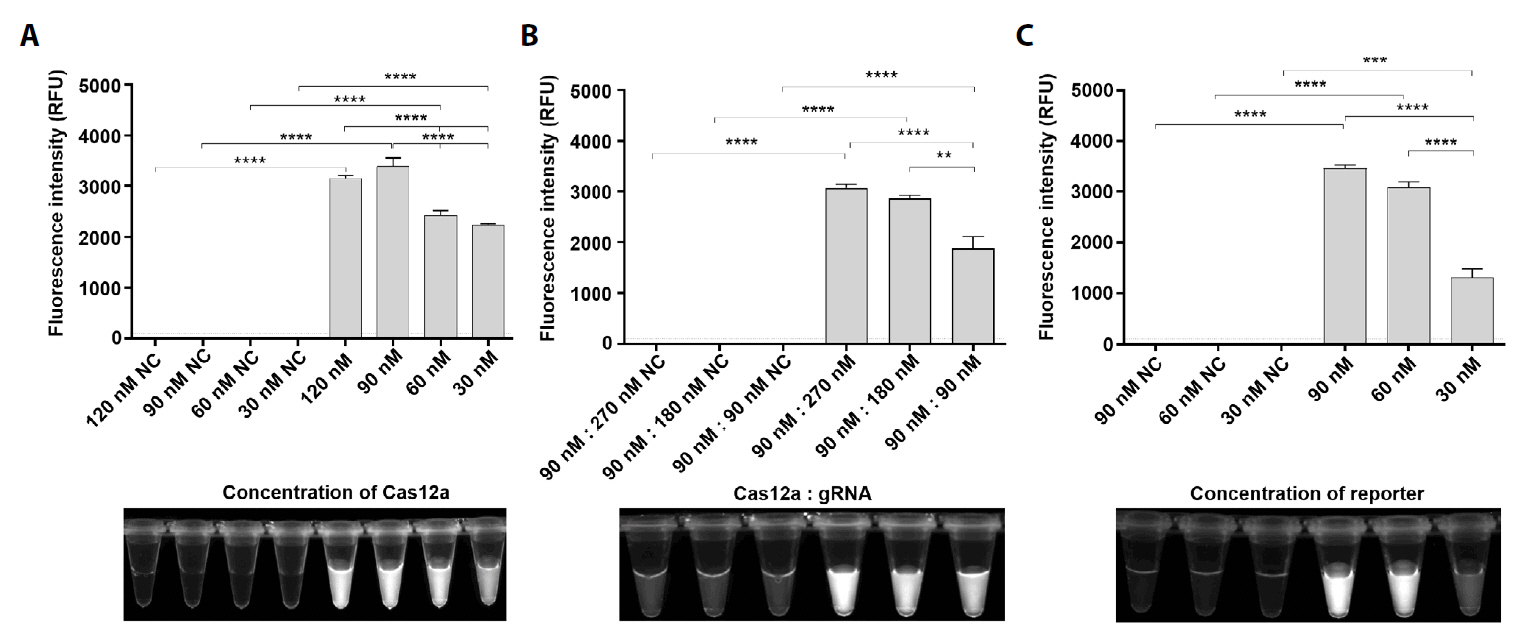
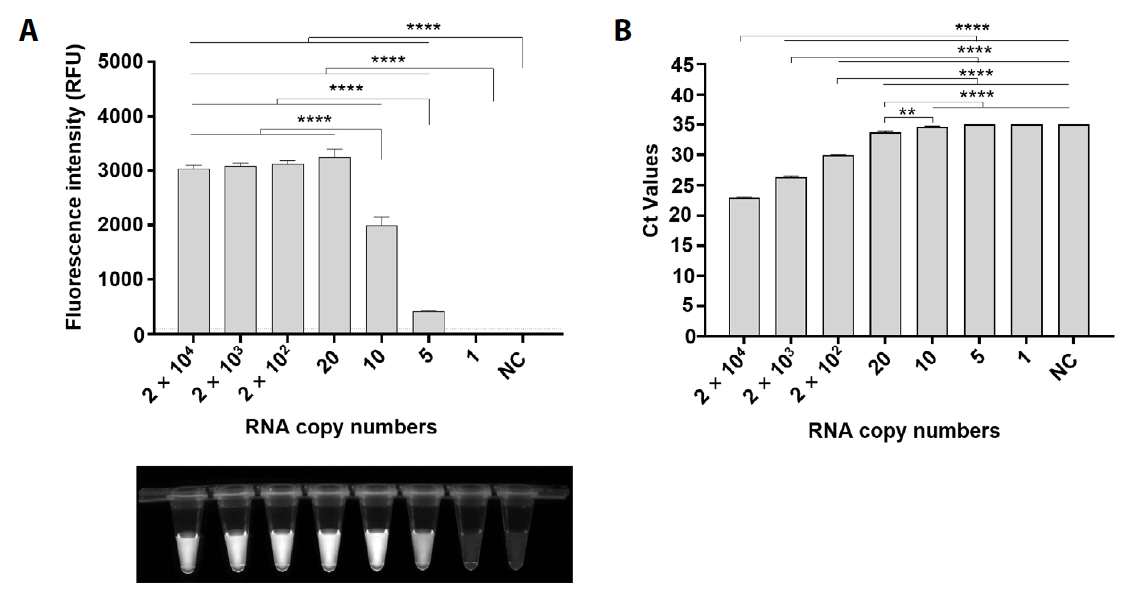
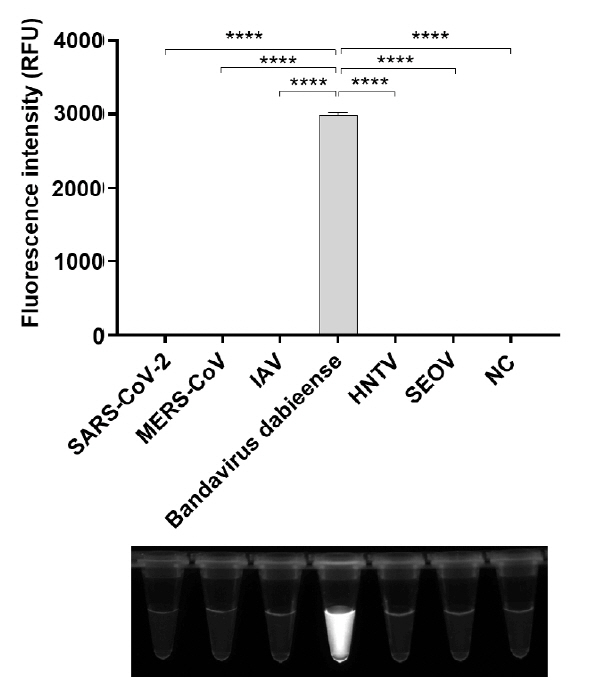
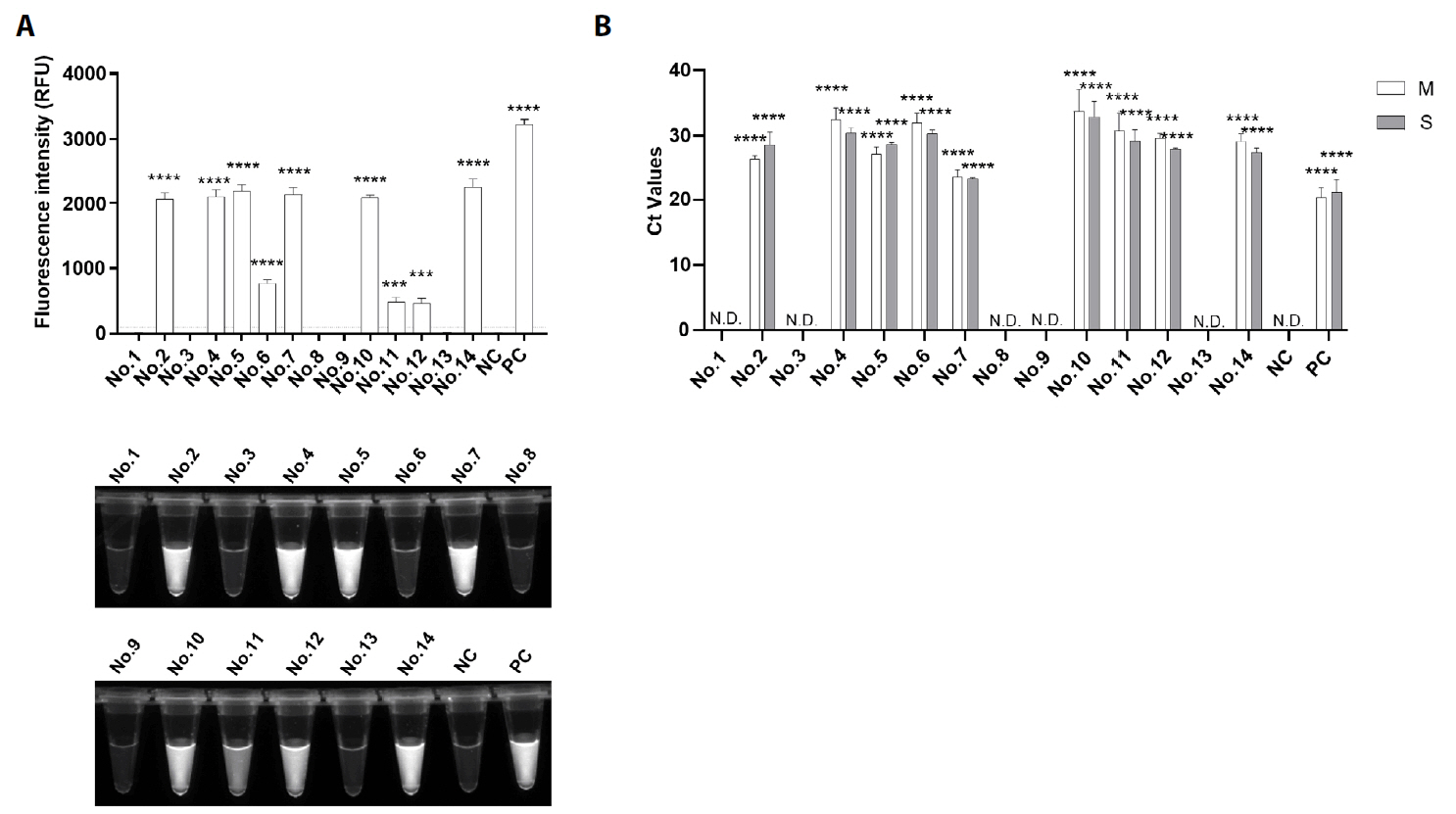
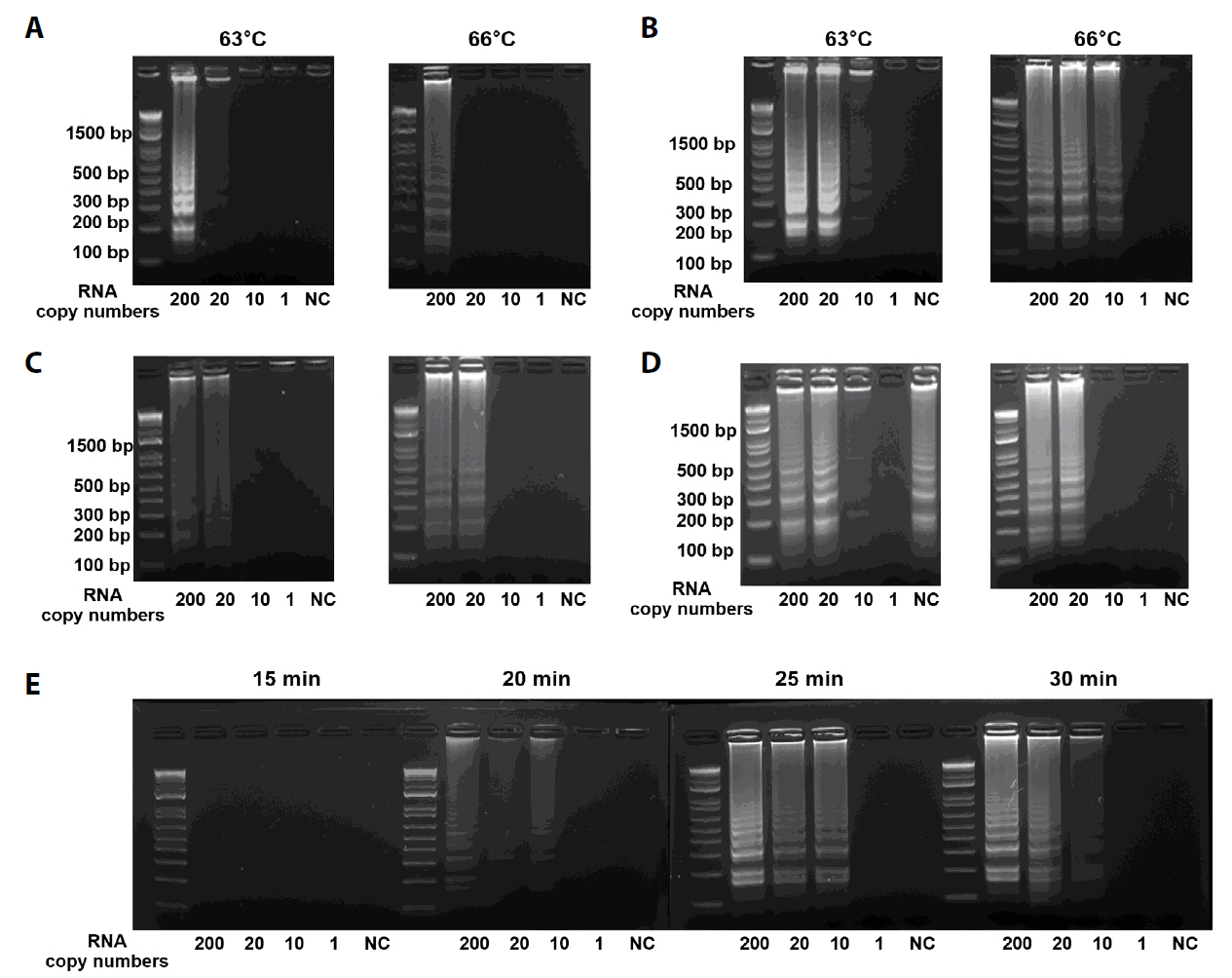
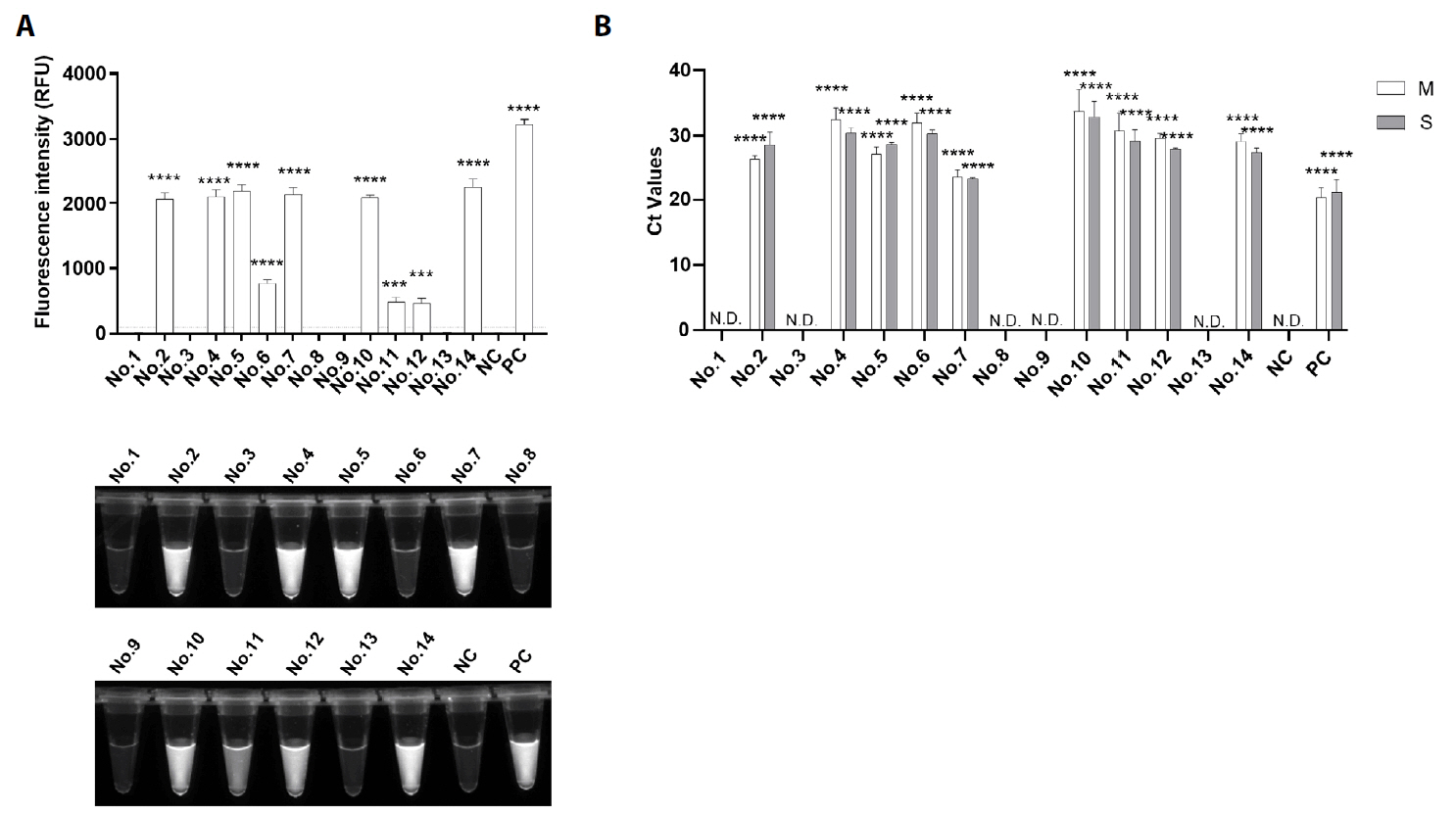
Fig. 1.
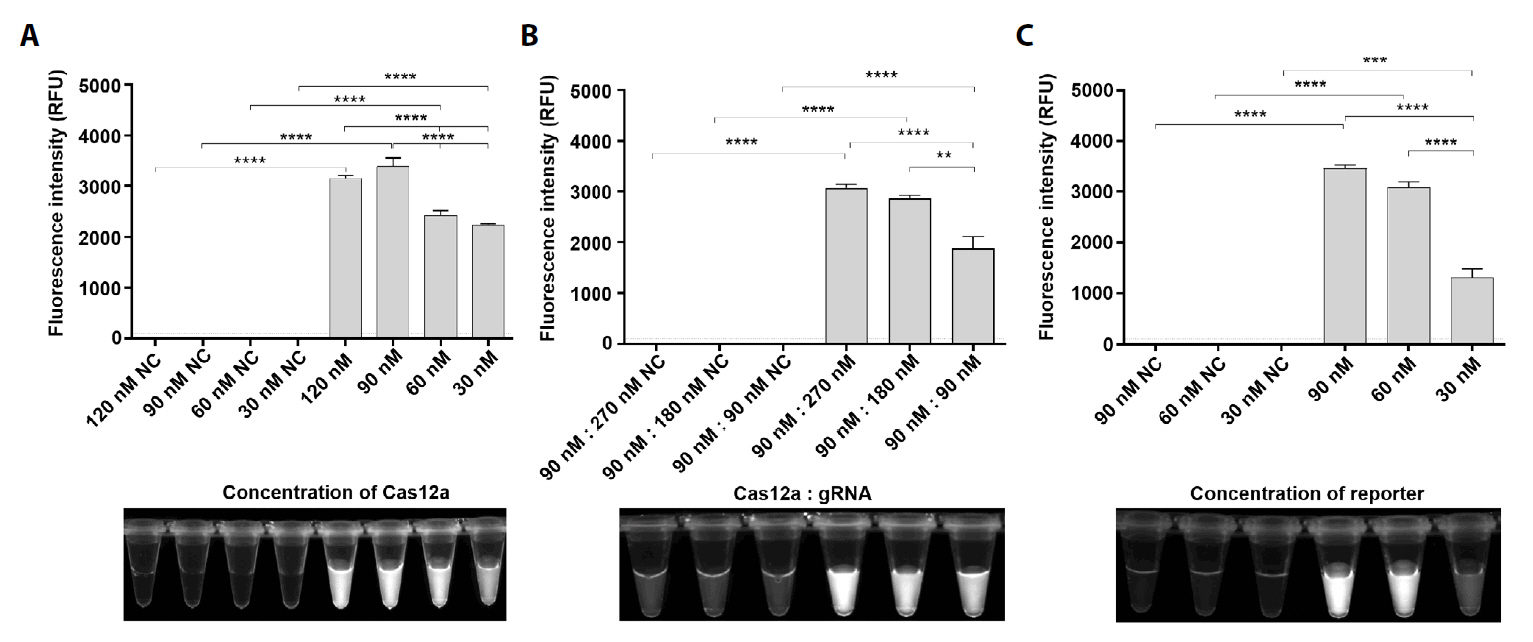
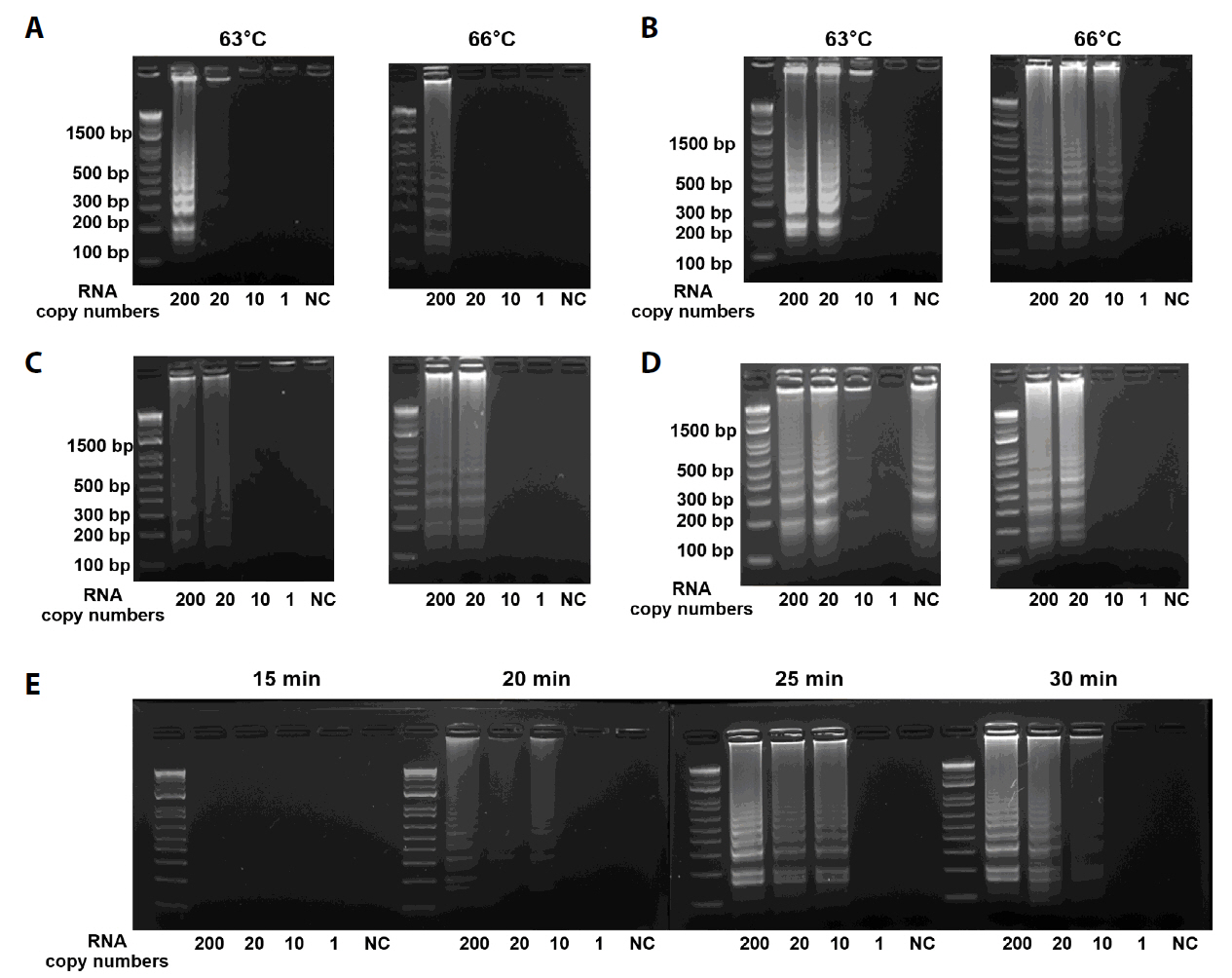
Fig. 2.
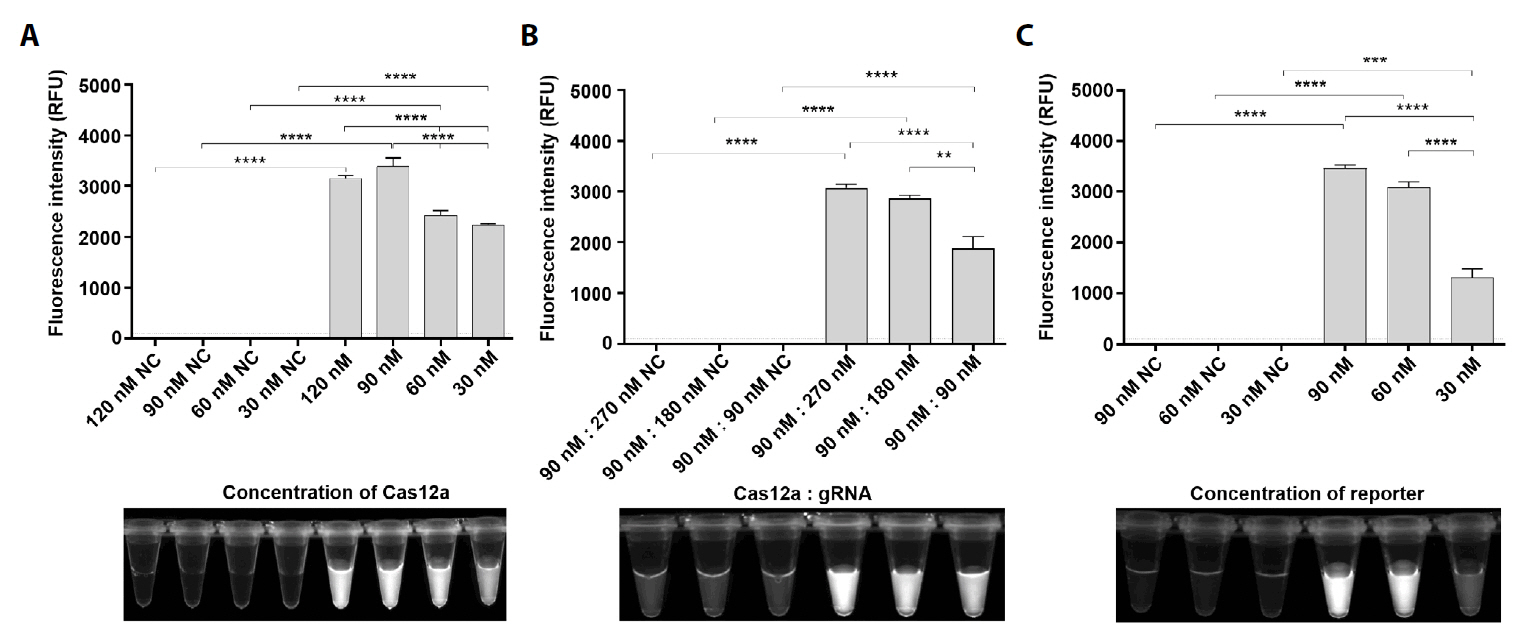
Fig. 3.
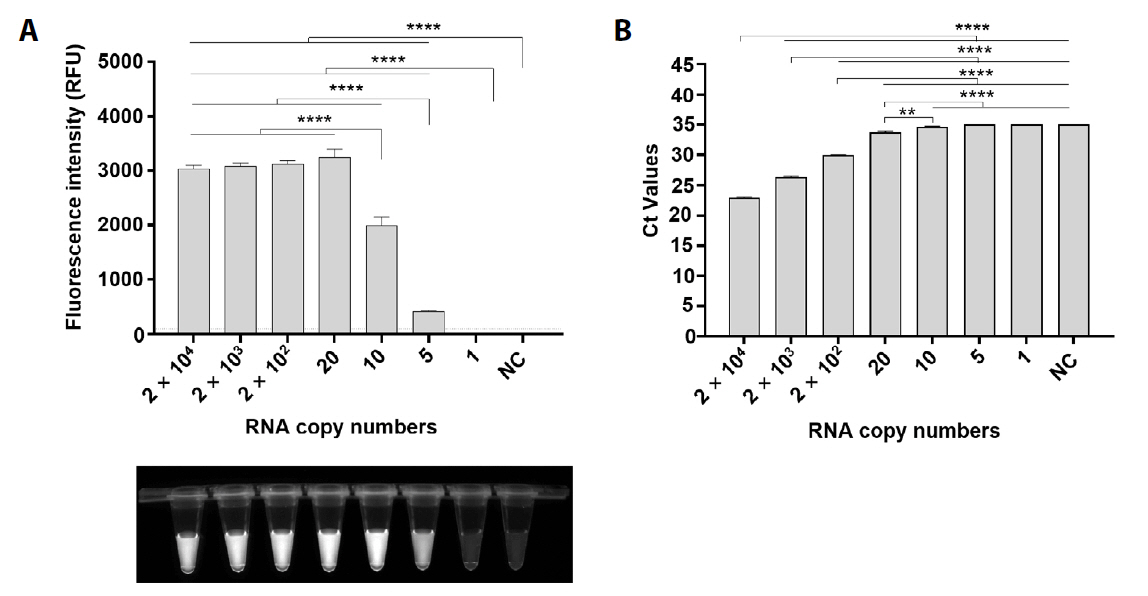
Fig. 4.
Fig. 5.
| Primer | Sequences (5` → 3`) | Position | |
|---|---|---|---|
| RT-LAMP | F3 | GGTGCATGCGAATCTGTCT | 2727–2745 |
| B3 | AGCCAGGGCCAAGACTYC | 2928–2945 | |
| FIP | TGCACTAGCCGGGCATTAGAATCAGCATGGTGG | 2798–2814/2754–2769 | |
| BIP | CATGAGACTGTTGCCAACCCTCTGACTTCAGCCCATGGTT | 2861–2881/2906–2924 | |
| LF | GCATCCATAACGTAGAT | 2776–2792 | |
| LB | GGCTYAAGAATTCYATCATAGA | 2883–2905 | |
| gRNA | F | CCCTAATACGACTCACTATAGGTAATTTCTACTAAGTGTAGAT | - |
| R | ACATCTAGCCATGGTCTCAACCCATCTACACTTAGTAGAAATTA | - | |
| FQ reporter | FAM-TTATT-BHQ1 | - | |
| qRT-PCR | F | ATTCAAGCCTGCCTTAAGTTCAAG | 1246–1270 |
| R | TTTCTTCTGGTTTGCTGCCATT | 1321–1342 | |
| F3 (µM) | B3 (µM) | FIP (µM) | BIP (µM) | LF (µM) | LB (µM) | |
|---|---|---|---|---|---|---|
| A | 0.08 | 0.1 | 0.86 | 0.86 | 0.14 | 0.17 |
| B | 0.18 | 0.2 | 0.86 | 0.86 | 0.14 | 0.17 |
| C | 0.18 | 0.2 | 0.74 | 0.74 | 0.14 | 0.17 |
| D | 0.18 | 0.2 | 0.86 | 0.86 | 0.04 | 0.07 |
Mixed base Y = C, T; Underlined, T7 promoter sequence; bold, LbCas12a mature direct repeat; italics, target nucleotide sequences (GGGTTGAGACCATGGCTAGATGT)
Table 1.
Table 2.
TOP
 MSK
MSK

 ePub Link
ePub Link Cite this Article
Cite this Article